![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | PiuraChilensis_Valdivia.fastq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 683347 |
| Filtered Sequences | 0 |
| Sequence length | 51-1201 |
| %GC | 44 |
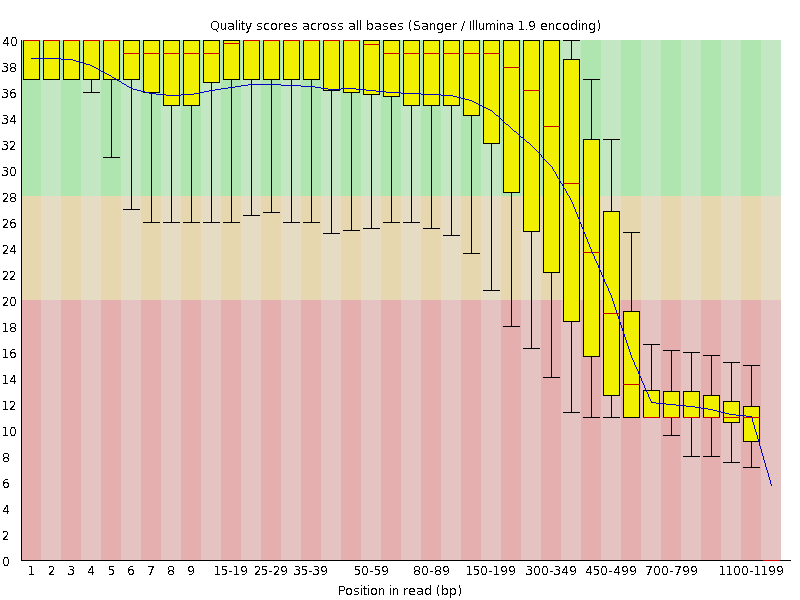
![[FAIL]](Icons/error.png) Per base sequence quality
Per base sequence quality
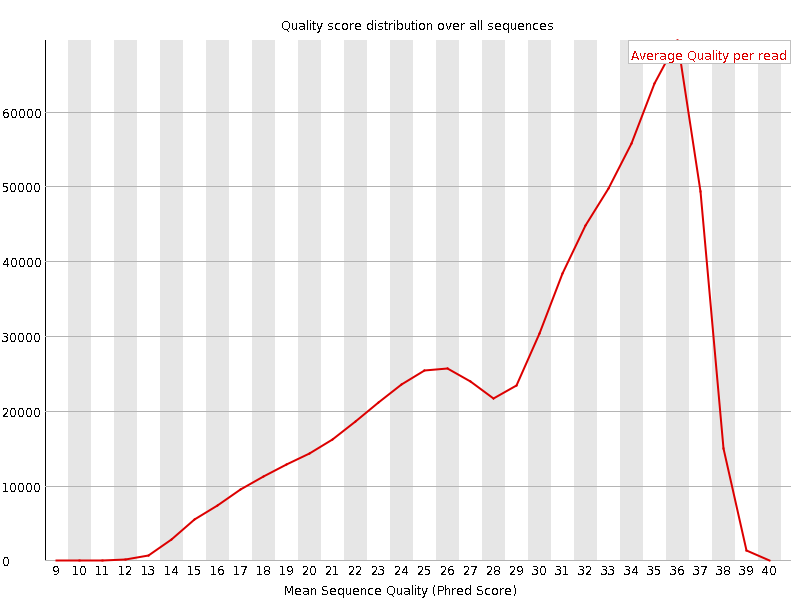
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
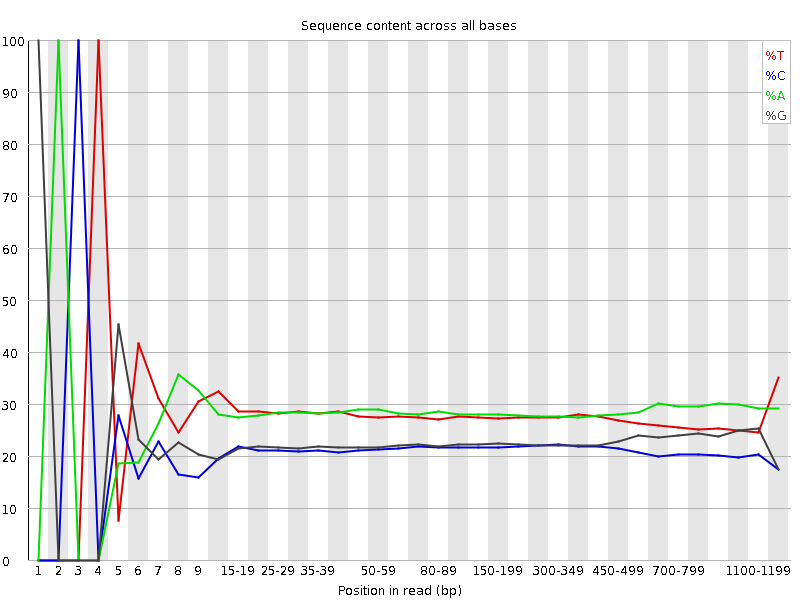
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
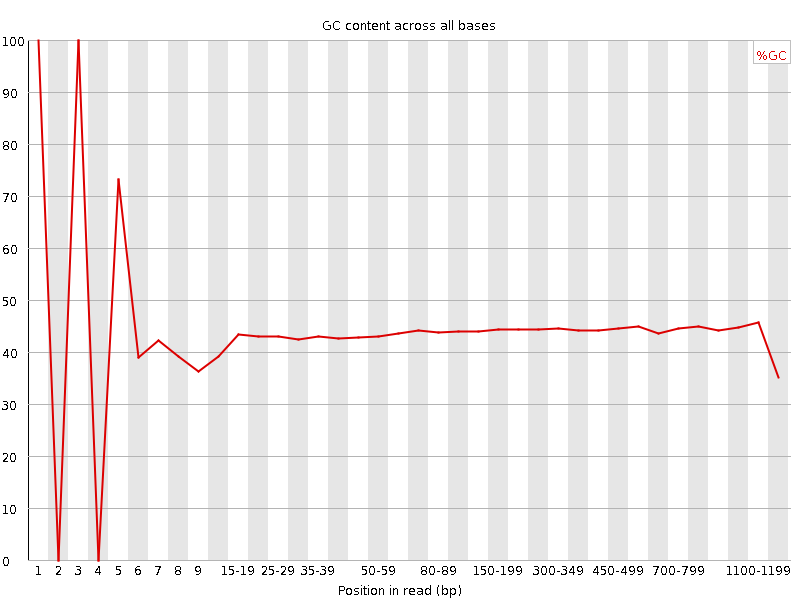
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
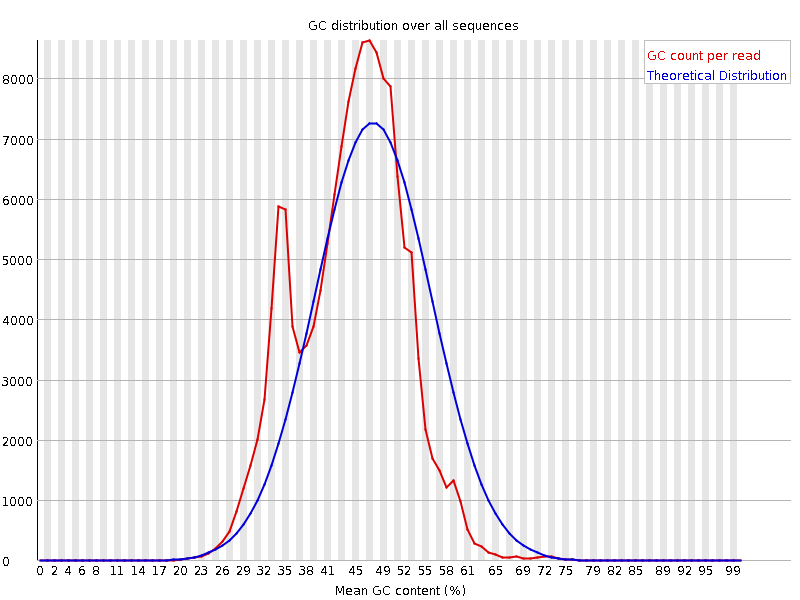
![[WARN]](Icons/warning.png) Per base N content
Per base N content
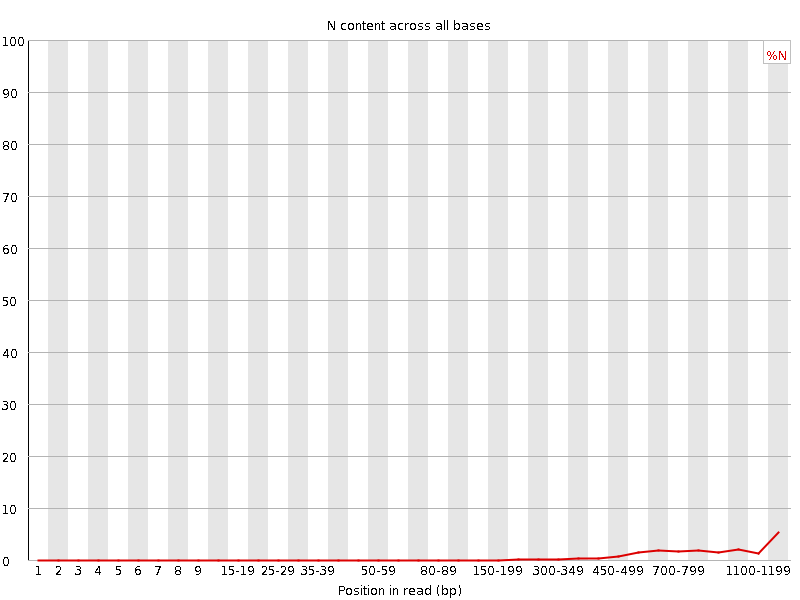
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
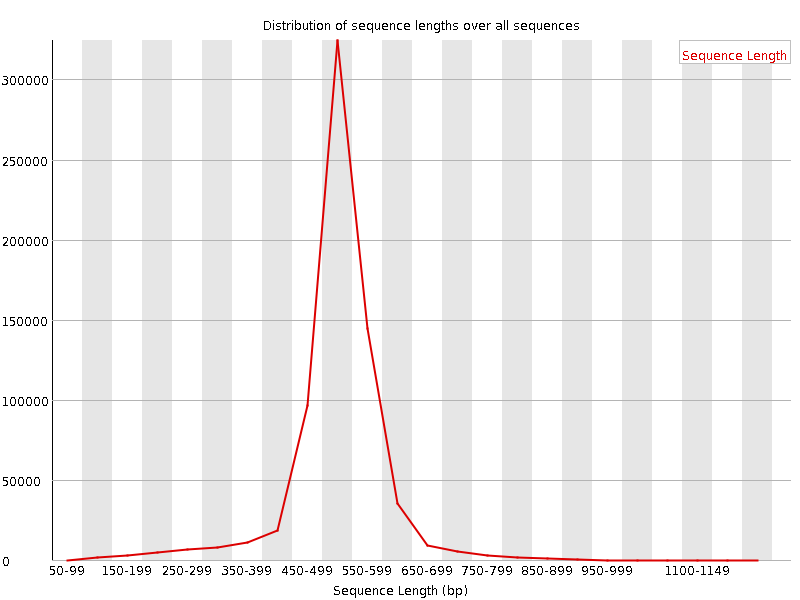
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GACTCGCTTGAAAAAATTCCTTTCGTACTATTTCTCTGTTTTTTTAAAAG | 1200 | 0.1756062439726815 | No Hit |
| GACTGCTTGAAAAAATTCCTTTCGTACTATTTCTCTGTTTTTTTAAAAGA | 1007 | 0.14736290640040858 | No Hit |
| GACTCCGCTTGAAAAAATTCCTTTCGTACTATTTCTCTGTTTTTTTAAAA | 821 | 0.12014393858464294 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
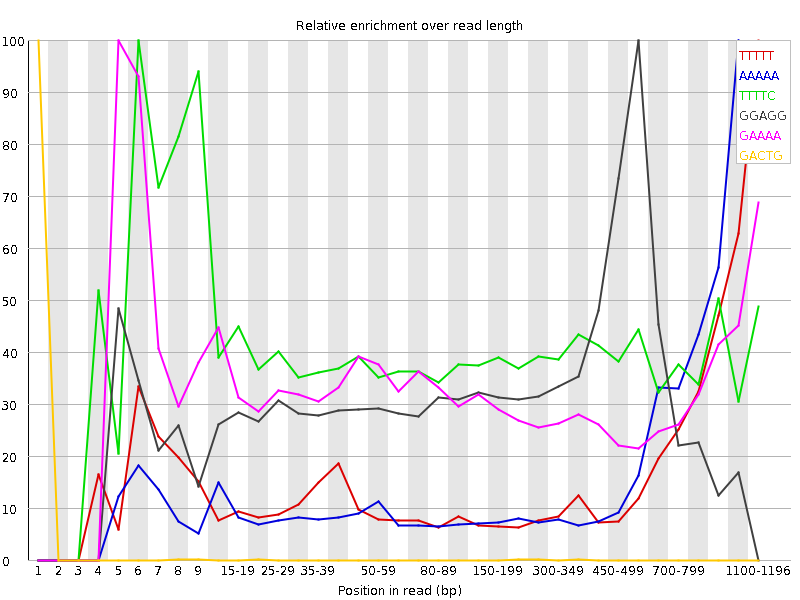
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 1490695 | 2.6458693 | 30.530987 | 1100-1196 |
| AAAAA | 1556275 | 2.5260768 | 29.264816 | 1000-1099 |
| TTTTC | 963285 | 2.164534 | 5.4971275 | 6 |
| GGAGG | 549795 | 2.1591082 | 5.315329 | 500-599 |
| GAAAA | 966195 | 1.9559301 | 6.94065 | 5 |
| GACTG | 590165 | 1.9550062 | 530.59985 | 1 |
| TCTTC | 685715 | 1.9506686 | 9.195696 | 4 |
| AGGCA | 590015 | 1.9198791 | 6.493845 | 500-599 |
| ATTTT | 1081130 | 1.884923 | 7.0113273 | 5 |
| GGGAG | 479640 | 1.8836014 | 5.4555254 | 500-599 |
| GTCGT | 537425 | 1.8124101 | 9.068862 | 1100-1196 |
| AAAAT | 1083145 | 1.7898256 | 5.5931735 | 7 |
| CGTGG | 428720 | 1.7712376 | 5.332869 | 500-599 |
| GTGGG | 422465 | 1.6889943 | 5.223289 | 500-599 |
| GGGGA | 412050 | 1.6181675 | 9.479918 | 1100-1196 |
| TGGGA | 496930 | 1.5929581 | 12.692512 | 4 |
| TGAAA | 763160 | 1.5727801 | 14.306586 | 4 |
| TCTTT | 682680 | 1.5340053 | 11.66585 | 4 |
| CTTTT | 679995 | 1.527972 | 15.62193 | 3 |
| CTTTC | 533875 | 1.518726 | 9.276291 | 3 |
| CTGGA | 450890 | 1.4936379 | 44.76265 | 3 |
| GACTC | 429240 | 1.4694016 | 334.87308 | 1 |
| AGGGG | 372905 | 1.4644407 | 8.672148 | 1100-1196 |
| GGAAA | 577935 | 1.4591364 | 7.1788077 | 5 |
| GTTTT | 670665 | 1.4583094 | 13.52818 | 1100-1196 |
| TCTTG | 527675 | 1.452582 | 12.893663 | 4 |
| TGGAA | 564975 | 1.452145 | 16.670115 | 4 |
| AAAAC | 692725 | 1.4491557 | 13.620111 | 1000-1099 |
| TGCTG | 429490 | 1.4484105 | 7.0068192 | 4 |
| GTACG | 435340 | 1.4421264 | 10.106037 | 600-699 |
| ACTTT | 650985 | 1.4368677 | 30.706446 | 2 |
| TCTCC | 397395 | 1.4311733 | 6.9162927 | 4 |
| CTCCA | 402980 | 1.425573 | 14.581375 | 3 |
| CGTAC | 415095 | 1.4209796 | 8.891791 | 600-699 |
| CTTGG | 417245 | 1.4071156 | 5.6853924 | 3 |
| TATTT | 805970 | 1.4051884 | 13.487421 | 4 |
| TGAAG | 546290 | 1.4041193 | 6.168473 | 4 |
| TCAAA | 651410 | 1.3873067 | 8.427012 | 4 |
| CTTTG | 503630 | 1.386391 | 5.7974935 | 3 |
| CTGCT | 397545 | 1.3854488 | 29.852356 | 3 |
| AGGAA | 544130 | 1.3737874 | 10.106939 | 1100-1196 |
| AAAAG | 673310 | 1.3630245 | 6.382084 | 1100-1196 |
| TCTGG | 402855 | 1.3585868 | 9.319243 | 4 |
| ACTTC | 484685 | 1.3543643 | 7.9519987 | 2 |
| CTGCC | 305465 | 1.3477077 | 36.985275 | 3 |
| CTGCA | 391020 | 1.3385645 | 25.467173 | 3 |
| ACTGG | 401355 | 1.3295462 | 125.11317 | 2 |
| TTTTA | 757240 | 1.3202288 | 10.438269 | 1100-1196 |
| TCGAC | 382945 | 1.3109217 | 6.2035875 | 4 |
| CTCTC | 363680 | 1.3097525 | 33.244377 | 3 |
| GTTGG | 400665 | 1.3075382 | 5.5355253 | 5 |
| TGTTG | 490630 | 1.306961 | 20.882004 | 4 |
| TGGAG | 407220 | 1.3053837 | 8.345987 | 4 |
| GTGAA | 504635 | 1.2970542 | 7.202284 | 5 |
| TGCAG | 391360 | 1.2964363 | 6.1653705 | 4 |
| TGTTT | 588480 | 1.2796044 | 25.643799 | 4 |
| GTTTC | 462025 | 1.2718608 | 5.9396315 | 5 |
| CTTGA | 467875 | 1.2651445 | 9.068419 | 3 |
| CTCCC | 277055 | 1.2631819 | 24.351948 | 3 |
| TAAAA | 759680 | 1.2553209 | 11.255014 | 1000-1099 |
| TCTGC | 359845 | 1.254064 | 6.144798 | 4 |
| CTGGG | 303530 | 1.2540207 | 37.28544 | 3 |
| CCCGG | 231490 | 1.2512124 | 28.400204 | 1100-1196 |
| CTCTG | 358950 | 1.2509449 | 28.468887 | 3 |
| ACGTA | 470110 | 1.2486649 | 9.339389 | 600-699 |
| GGTCG | 299355 | 1.2367718 | 16.538872 | 1100-1196 |
| ACTGC | 361195 | 1.2364658 | 98.03604 | 2 |
| GGGGG | 251775 | 1.2331461 | 62.048023 | 1100-1196 |
| GGGAA | 390980 | 1.2311182 | 7.953136 | 1100-1196 |
| AAATA | 744465 | 1.2301791 | 13.229909 | 1100-1196 |
| CCGGG | 233810 | 1.2229149 | 30.39504 | 1100-1196 |
| TGGTT | 457155 | 1.2177889 | 5.9329333 | 4 |
| CTCCT | 337240 | 1.2145319 | 12.244997 | 3 |
| CCCCG | 217430 | 1.2144619 | 28.193718 | 1100-1196 |
| TGTTC | 440830 | 1.2135153 | 16.271418 | 4 |
| TGCAA | 454590 | 1.2074419 | 8.681862 | 4 |
| TACGT | 445715 | 1.2052233 | 8.618544 | 600-699 |
| GTTTG | 451885 | 1.2037505 | 6.8670225 | 5 |
| ACTGT | 444685 | 1.2024382 | 184.99454 | 2 |
| GTCGA | 361575 | 1.1977692 | 5.4567685 | 5 |
| CTCTT | 420590 | 1.1964613 | 43.40424 | 3 |
| AAACC | 442595 | 1.1933136 | 19.174301 | 1000-1099 |
| CTGGT | 353820 | 1.1932212 | 20.855131 | 3 |
| GTTGT | 446505 | 1.1894189 | 7.883342 | 5 |
| TGCCC | 269565 | 1.1893175 | 7.2560825 | 4 |
| CTGTT | 430610 | 1.1853819 | 84.58521 | 3 |
| ACTTG | 431865 | 1.1677727 | 21.830484 | 2 |
| TCGAA | 439510 | 1.1673877 | 7.921857 | 4 |
| AAGGA | 460545 | 1.1627569 | 20.213879 | 1100-1196 |
| ACCGG | 277185 | 1.1624507 | 19.009424 | 1100-1196 |
| CTGTC | 331980 | 1.1569542 | 56.82998 | 3 |
| CTTGC | 331850 | 1.1565012 | 5.992032 | 3 |
| TCCGG | 270720 | 1.1558168 | 6.515348 | 4 |
| CTGTG | 341700 | 1.1523479 | 43.09249 | 3 |
| CTGAA | 432595 | 1.1490208 | 34.501247 | 3 |
| CTCCG | 259590 | 1.145308 | 26.431055 | 3 |
| AACGT | 429630 | 1.1411453 | 9.950807 | 700-799 |
| TGTCG | 338075 | 1.1401228 | 16.647713 | 4 |
| TCGCC | 258390 | 1.1400135 | 5.9140487 | 4 |
| CGAAC | 338340 | 1.1377053 | 26.922117 | 1100-1196 |
| CCTTT | 399505 | 1.1364805 | 5.413853 | 1000-1099 |
| CCGGT | 264960 | 1.131225 | 16.359179 | 1100-1196 |
| GGTTT | 422930 | 1.1266189 | 17.937471 | 1100-1196 |
| TGCTT | 406840 | 1.1199479 | 9.849424 | 4 |
| GCTTG | 331220 | 1.1170051 | 5.2074537 | 5 |
| CTCGA | 325965 | 1.1158642 | 27.920351 | 3 |
| TGGAC | 336710 | 1.1154002 | 6.575256 | 4 |
| ACGTT | 412120 | 1.1143818 | 9.437982 | 700-799 |
| TGGAT | 425020 | 1.112126 | 6.380915 | 4 |
| TGTGG | 339625 | 1.1083391 | 10.784704 | 4 |
| CCCTT | 306715 | 1.1045996 | 5.410959 | 1000-1099 |
| AACCC | 317240 | 1.1023766 | 20.193499 | 1100-1196 |
| TGCTC | 315020 | 1.0978484 | 5.273386 | 4 |
| GACTT | 405735 | 1.0971165 | 74.41268 | 1 |
| TGTGA | 418605 | 1.0953401 | 16.566097 | 4 |
| TGTCA | 403935 | 1.0922492 | 16.415283 | 4 |
| ACGGA | 335665 | 1.0922371 | 6.5879016 | 1100-1196 |
| CGGGG | 215510 | 1.0907745 | 32.268963 | 1100-1196 |
| AACGG | 334980 | 1.090008 | 20.24446 | 1100-1196 |
| GTCAA | 408300 | 1.0844905 | 6.1897736 | 5 |
| TGTCC | 310980 | 1.0837687 | 8.821916 | 4 |
| GTTGA | 414025 | 1.0833559 | 5.4569893 | 5 |
| TGGCA | 326955 | 1.0830854 | 11.929394 | 4 |
| CTGGC | 253535 | 1.0824469 | 38.62957 | 3 |
| GTTCA | 399075 | 1.0791076 | 5.4579797 | 5 |
| TGCCA | 314150 | 1.0754182 | 10.942316 | 4 |
| CCGTT | 305860 | 1.0659257 | 13.03819 | 1100-1196 |
| TTTAC | 481375 | 1.062501 | 7.140416 | 800-899 |
| GGTAC | 320300 | 1.0610397 | 6.9663653 | 700-799 |
| TCGAG | 319990 | 1.0600128 | 5.132116 | 4 |
| TCCGA | 309185 | 1.0584217 | 6.547741 | 4 |
| AAGGG | 335285 | 1.0557457 | 8.760144 | 1100-1196 |
| AAACG | 404320 | 1.0548913 | 11.095908 | 1100-1196 |
| TCCCC | 230755 | 1.0520855 | 6.4835305 | 1100-1196 |
| GTACC | 306160 | 1.0480664 | 6.05285 | 700-799 |
| CCCCC | 181470 | 1.0474538 | 60.887226 | 1100-1196 |
| ACTCC | 295825 | 1.0465039 | 66.69485 | 2 |
| GTAAC | 391780 | 1.0406115 | 10.105322 | 1100-1196 |
| TTACC | 371670 | 1.0385642 | 9.240573 | 800-899 |
| GTTAC | 383425 | 1.0367897 | 8.017845 | 700-799 |
| CTCGT | 297475 | 1.0367038 | 14.355722 | 3 |
| TGGGG | 258570 | 1.0337502 | 11.047894 | 4 |
| TCGGA | 311690 | 1.0325179 | 7.429186 | 4 |
| ACTGA | 388595 | 1.0321519 | 67.3436 | 2 |
| GACCC | 237950 | 1.0312314 | 34.697903 | 1100-1196 |
| AACCG | 306660 | 1.0311779 | 18.861559 | 1000-1099 |
| GACTA | 388005 | 1.0305847 | 177.32678 | 1 |
| TCCCA | 291080 | 1.029718 | 5.881836 | 4 |
| ACTAC | 374305 | 1.0273955 | 15.629284 | 2 |
| CGTTT | 372215 | 1.0246322 | 10.878089 | 1100-1196 |
| TCTCG | 293180 | 1.0217357 | 10.079624 | 4 |
| AATAA | 616590 | 1.0188742 | 6.6149545 | 1100-1196 |
| TCCCG | 230485 | 1.0168971 | 7.2560835 | 4 |
| TGGCC | 237855 | 1.0155025 | 5.37076 | 4 |
| GGGTT | 310715 | 1.0139936 | 21.940968 | 1100-1196 |
| CTGAC | 295850 | 1.0127726 | 12.230597 | 3 |
| GTAAA | 486765 | 1.0031636 | 6.9713755 | 900-999 |
| TCTGA | 370370 | 1.0014888 | 6.7473445 | 4 |
| ACGGT | 301950 | 1.0002527 | 12.383497 | 800-899 |
| ACCGT | 291885 | 0.9991993 | 9.933352 | 800-899 |
| CCCGT | 225590 | 0.99530035 | 9.66037 | 1100-1196 |
| GGTAA | 384495 | 0.98826057 | 16.292736 | 1100-1196 |
| ACCCC | 220350 | 0.9868454 | 23.36164 | 1100-1196 |
| TCGGG | 238625 | 0.9858686 | 5.3356586 | 4 |
| GAACT | 370385 | 0.983784 | 5.3775268 | 1100-1196 |
| TCAGA | 370155 | 0.9831732 | 5.0393147 | 4 |
| TCTCA | 349885 | 0.9776901 | 7.484084 | 4 |
| TGGCT | 289880 | 0.9775902 | 6.337433 | 4 |
| ACCCG | 224800 | 0.9742418 | 12.805767 | 1000-1099 |
| ACGGG | 238935 | 0.9696589 | 18.00665 | 1100-1196 |
| TTTAA | 565145 | 0.96785814 | 14.752424 | 1000-1099 |
| GGGGT | 241655 | 0.9661248 | 26.87944 | 1100-1196 |
| ACTCT | 345635 | 0.96581423 | 99.90418 | 2 |
| TGTCT | 350650 | 0.9652681 | 7.62124 | 4 |
| TTAAA | 570565 | 0.9598272 | 14.238287 | 1000-1099 |
| TTACG | 353170 | 0.95497954 | 7.769322 | 700-799 |
| CTCAA | 347125 | 0.9527916 | 22.754753 | 3 |
| TGGCG | 230460 | 0.9521352 | 9.542416 | 4 |
| CGGTT | 281950 | 0.95084715 | 13.586904 | 900-999 |
| CTCGC | 215150 | 0.94923913 | 28.682924 | 3 |
| TCGTA | 348435 | 0.94217604 | 5.474526 | 1100-1196 |
| TACCG | 274315 | 0.9390525 | 9.109239 | 700-799 |
| CTGAT | 346915 | 0.9380658 | 14.791072 | 3 |
| CGGTA | 282365 | 0.9353747 | 13.192894 | 1100-1196 |
| CGTAA | 350495 | 0.930954 | 7.765349 | 700-799 |
| TATTC | 418430 | 0.9235675 | 5.501986 | 4 |
| CTCGG | 216255 | 0.923283 | 23.023664 | 3 |
| ACTCG | 269170 | 0.9214398 | 82.735 | 2 |
| CTATT | 413785 | 0.91331494 | 35.50377 | 3 |
| TACCC | 255560 | 0.90406334 | 7.104487 | 1100-1196 |
| GGGTA | 280940 | 0.9005809 | 10.603396 | 1100-1196 |
| TACGG | 270995 | 0.89770997 | 9.054101 | 700-799 |
| CGGGT | 216355 | 0.89386106 | 13.925174 | 1100-1196 |
| TTAAC | 409370 | 0.88756037 | 10.079556 | 1100-1196 |
| TGCCT | 254410 | 0.8866218 | 5.974108 | 4 |
| CTCAC | 248635 | 0.8795656 | 12.511349 | 3 |
| TAAAC | 409950 | 0.87306964 | 9.698237 | 1100-1196 |
| TAACC | 317950 | 0.87271166 | 13.294551 | 1100-1196 |
| GTTAA | 409170 | 0.85846 | 10.605812 | 900-999 |
| TCGCT | 246065 | 0.85753936 | 8.103227 | 4 |
| CTCAG | 249720 | 0.8548574 | 14.666126 | 3 |
| CCGTA | 248715 | 0.85141706 | 7.1465573 | 700-799 |
| CTCAT | 302050 | 0.8440239 | 8.211544 | 3 |
| TAACG | 317555 | 0.84346163 | 10.334424 | 700-799 |
| TATTA | 487465 | 0.8348246 | 6.0304503 | 4 |
| ACTCA | 303140 | 0.8320612 | 52.286285 | 2 |
| TATCA | 382855 | 0.8300729 | 5.510691 | 4 |
| GTTTA | 388270 | 0.82930446 | 9.199447 | 1000-1099 |
| TAGAA | 401895 | 0.82825685 | 6.6618857 | 4 |
| GGTTA | 315585 | 0.82577354 | 10.542085 | 900-999 |
| CTGTA | 304740 | 0.8240238 | 23.267014 | 3 |
| CGTTA | 302565 | 0.81814253 | 9.195375 | 700-799 |
| TGTGC | 241680 | 0.81504065 | 5.2507668 | 4 |
| CCCCT | 176695 | 0.8056088 | 6.3098655 | 1100-1196 |
| TCCGC | 181315 | 0.79995954 | 6.3917217 | 4 |
| TCGCA | 233395 | 0.7989726 | 6.035923 | 4 |
| CTGAG | 241130 | 0.7987778 | 10.98999 | 3 |
| CTACG | 233210 | 0.7983393 | 5.329928 | 3 |
| TGCGA | 238550 | 0.79023117 | 6.122674 | 4 |
| ACTAA | 369285 | 0.7864656 | 42.940514 | 2 |
| TGCCG | 181855 | 0.776415 | 7.80301 | 4 |
| CTGCG | 180840 | 0.77208155 | 18.148193 | 3 |
| ACTAG | 290610 | 0.7718928 | 31.124846 | 2 |
| GAGTG | 240695 | 0.77157146 | 10.553847 | 1000-1099 |
| CTAGT | 280075 | 0.7573291 | 5.478692 | 3 |
| ACTAT | 347800 | 0.75406975 | 63.275627 | 2 |
| TGTAA | 350210 | 0.73475885 | 7.5229926 | 4 |
| CTAAA | 344640 | 0.73397917 | 21.866154 | 3 |
| TGTTA | 330445 | 0.70579624 | 11.072342 | 4 |
| ACTTA | 302955 | 0.6568407 | 5.8290477 | 2 |
| CTAGG | 197200 | 0.6532534 | 5.3370204 | 3 |
| CTAAC | 236580 | 0.64936674 | 6.7358594 | 3 |
| CGCTT | 185675 | 0.64707947 | 5.075895 | 5 |
| CTATC | 229640 | 0.6416873 | 17.21543 | 3 |
| ATAAC | 290035 | 0.6176869 | 5.727066 | 1100-1196 |
| TATCG | 216985 | 0.58673227 | 5.0744486 | 4 |
| CTATG | 210685 | 0.56969696 | 9.549374 | 3 |
| CTCTA | 199560 | 0.5576342 | 8.650933 | 3 |
| CTAAT | 246455 | 0.53434235 | 9.305511 | 3 |
| CTAGA | 200400 | 0.5322849 | 14.71387 | 3 |
| CTAAG | 173030 | 0.4595871 | 8.366844 | 3 |
| CTATA | 175825 | 0.38120848 | 7.3885202 | 3 |
| CTAGC | 109700 | 0.37553203 | 8.700843 | 3 |