6.1 텍스트 파일 이용하는 방법¶
파이썬 좋은 이유¶
- 단순한 문법
- 직관적인 자료 구조
- 튜플에 데이터를 저장하고 읽어내는 편리한 기능
pandas 파일 파싱 함수¶
| 함수 | 설명 |
|---|---|
| read_csv | 파일, URL 또는 파일과 유사한 객체로부터 구분된 데이터를 읽어온다. 데이터 구분자는 쉼표(,)를 기본으로 한다. |
| read_table | 파일, URL 또는 파일과 유사한 객체로부터 구분된 데이터를 읽어온다. 데이터 구분자는 탭('\t')을 기본으로 한다. |
| read_fwf | 고정폭 칼럼 형식에서 데이터를 읽어온다(구분자가 없는 데이터) |
| read_clipboard | 클립보드에 있는 데이터를 읽어오는 read_table 함수. 웹페이지에서 표를 긁어올 때 유용하다. |
pandas 파일 파싱 함수 옵션¶
- 색인: 반환하는 DataFrame에서 하나 이상의 칼럼을 색인으로 지정할 수 있다. 파일이나 사용자로부터 칼럼의 이름을 받거나 아무것도 받지 않을 수 있다.
- 자료형 추론과 데이터 변환: 사용자 정의 값 변환과 비어있는 값을 위한 사용자 리스트를 포함한다.
- 날짜 분석: 여러 칼럼에 걸쳐 있는 날짜와 시간 정보를 하나의 칼럼에 조합해서 결과에 반영한다.
- 반복: 여러 파일에 걸쳐 있는 자료를 반복적으로 읽어올 수 있다.
- 정제되지 않는 데이터 처리: 로우나 꼬리말, 주석 건너뛰기 또는 천 단위마다 쉼표로 구분된 숫자 같은 사소한 일을 처리해준다.
자료형 추론은 매우 중요¶
- 어떤 칼럼이 숫자인지 불리언인지 지정해줄 필요가 없다
from pandas import DataFrame, Series
import pandas as pd
!cat ch06/ex1.csv
a,b,c,d,message 1,2,3,4,hello 5,6,7,8,world 9,10,11,12,foo
df = pd.read_csv('ch06/ex1.csv')
pd.read_csv('ch06/ex1.csv', header=None)
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | a | b | c | d | message |
| 1 | 1 | 2 | 3 | 4 | hello |
| 2 | 5 | 6 | 7 | 8 | world |
| 3 | 9 | 10 | 11 | 12 | foo |
# 원래 있던 Column명 무시하고 내가 원하는 Column명 설정
pd.read_csv('ch06/ex1.csv', names=[5,6,7,8,9])
| 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|
| 0 | a | b | c | d | message |
| 1 | 1 | 2 | 3 | 4 | hello |
| 2 | 5 | 6 | 7 | 8 | world |
| 3 | 9 | 10 | 11 | 12 | foo |
pd.read_csv('ch06/ex1.csv', names=['a1', 'b1', 'c1', 'd1', 'message1'])
| a1 | b1 | c1 | d1 | message1 | |
|---|---|---|---|---|---|
| 0 | a | b | c | d | message |
| 1 | 1 | 2 | 3 | 4 | hello |
| 2 | 5 | 6 | 7 | 8 | world |
| 3 | 9 | 10 | 11 | 12 | foo |
df
| a | b | c | d | message | |
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | hello |
| 1 | 5 | 6 | 7 | 8 | world |
| 2 | 9 | 10 | 11 | 12 | foo |
# csv는 DataFrame으로 읽어온다.
type(df)
pandas.core.frame.DataFrame
pd.read_table('ch06/ex1.csv', sep=',')
| a | b | c | d | message | |
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | hello |
| 1 | 5 | 6 | 7 | 8 | world |
| 2 | 9 | 10 | 11 | 12 | foo |
pd.read_table('ch06/ex1.csv', sep=',', header=None)
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | a | b | c | d | message |
| 1 | 1 | 2 | 3 | 4 | hello |
| 2 | 5 | 6 | 7 | 8 | world |
| 3 | 9 | 10 | 11 | 12 | foo |
!cat ch06/ex2.csv
1,2,3,4,hello 5,6,7,8,world 9,10,11,12,foo
# header 자동 생성
pd.read_csv('ch06/ex2.csv', header=None)
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | hello |
| 1 | 5 | 6 | 7 | 8 | world |
| 2 | 9 | 10 | 11 | 12 | foo |
# header 옵션이 없을시 header를 첫번째 줄로 이용
pd.read_csv('ch06/ex2.csv')
| 1 | 2 | 3 | 4 | hello | |
|---|---|---|---|---|---|
| 0 | 5 | 6 | 7 | 8 | world |
| 1 | 9 | 10 | 11 | 12 | foo |
# Column명 추가
pd.read_csv('ch06/ex2.csv', names=['a', 'b', 'c', 'message'])
| a | b | c | message | |
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | hello |
| 5 | 6 | 7 | 8 | world |
| 9 | 10 | 11 | 12 | foo |
names = ['a', 'b', 'c', 'd', 'message']
pd.read_csv('ch06/ex2.csv', names=names)
| a | b | c | d | message | |
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | hello |
| 1 | 5 | 6 | 7 | 8 | world |
| 2 | 9 | 10 | 11 | 12 | foo |
# message -> index
pd.read_csv('ch06/ex2.csv', names=names, index_col='message')
| a | b | c | d | |
|---|---|---|---|---|
| message | ||||
| hello | 1 | 2 | 3 | 4 |
| world | 5 | 6 | 7 | 8 |
| foo | 9 | 10 | 11 | 12 |
pd.read_csv('ch06/ex2.csv', names=names, index_col='a')
| b | c | d | message | |
|---|---|---|---|---|
| a | ||||
| 1 | 2 | 3 | 4 | hello |
| 5 | 6 | 7 | 8 | world |
| 9 | 10 | 11 | 12 | foo |
!cat ch06/csv_mindex.csv
key1,key2,value1,value2 one,a,1,2 one,b,3,4 one,c,5,6 one,d,7,8 two,a,9,10 two,b,11,12 two,c,13,14 two,d,15,16
계층적 색인을 지정하고 싶다면 칼럼 번호나 이름의 리스트를 넘긴다¶
- 2번째 공부하면서 정리하니 계층적 색인을 어떻게 사용하는지 조금은 이해가 간다.
parsed = pd.read_csv('ch06/csv_mindex.csv', index_col=['key1', 'key2'])
parsed
| value1 | value2 | ||
|---|---|---|---|
| key1 | key2 | ||
| one | a | 1 | 2 |
| b | 3 | 4 | |
| c | 5 | 6 | |
| d | 7 | 8 | |
| two | a | 9 | 10 |
| b | 11 | 12 | |
| c | 13 | 14 | |
| d | 15 | 16 |
고정된 구분자가 없다면 read_table의 구분자로 정규표현식을 사용하면 된다.¶
list(open('ch06/ex3.txt'))
[' A B C\n', 'aaa -0.264438 -1.026059 -0.619500\n', 'bbb 0.927272 0.302904 -0.032399\n', 'ccc -0.264273 -0.386314 -0.217601\n', 'ddd -0.871858 -0.348382 1.100491\n']
직접 파일을 고쳐도 되지만 이 파일은 여러 개의 공백문자로 필드가 구분되어 있으므로 이를 표현할 수 있는 정규표현식 \s+를 사용해서 처리¶
result = pd.read_table('ch06/ex3.txt', sep='\s+')
result
| A | B | C | |
|---|---|---|---|
| aaa | -0.264438 | -1.026059 | -0.619500 |
| bbb | 0.927272 | 0.302904 | -0.032399 |
| ccc | -0.264273 | -0.386314 | -0.217601 |
| ddd | -0.871858 | -0.348382 | 1.100491 |
이 경우, 첫번째 로우는 다른 로우보다 칼럼이 하나 적기 때문에 read_table은 첫 번째 칼럼이 DataFrame의 색인이 되어야 한다고 추론¶
pd.read_csv('ch06/ex3.txt', delimiter='\s+')
| A | B | C | |
|---|---|---|---|
| aaa | -0.264438 | -1.026059 | -0.619500 |
| bbb | 0.927272 | 0.302904 | -0.032399 |
| ccc | -0.264273 | -0.386314 | -0.217601 |
| ddd | -0.871858 | -0.348382 | 1.100491 |
IO Tools(Text, CSV, HDF5, ⋯) example¶
- 파서 함수는 파일 형식에서 발생할 수 있는 매우 다양한 예외를 잘 처리할 수 있도록 많은 추가 인자를 가지고 있다.
- skiprows를 이용해서 첫번째, 세번째, 네번째 로우를 건너뛸 수 있음
# Read CSV(comma-separated) file into DataFrame
pd.read_csv?
!cat ch06/ex4.csv
# hey! a,b,c,d,message # just wanted to make things more difficult for you # who reads CSV files with computers, anyway? 1,2,3,4,hello 5,6,7,8,world 9,10,11,12,foo
pd.read_csv('ch06/ex4.csv', skiprows=[0, 2, 3])
| a | b | c | d | message | |
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | hello |
| 1 | 5 | 6 | 7 | 8 | world |
| 2 | 9 | 10 | 11 | 12 | foo |
- 누락된 값을 잘 처리하는 일은 파일을 읽는 과정에서 자주 발생하는 일이고 중요한 문제
- 누락된 값은 표기하지 않거나(비어있는 문자열) 구분하기 쉬운 특수한 문자로 표기
- NA, -1, #IND, NULL처럼 비어있는 값으로 인식
!cat ch06/ex5.csv
something,a,b,c,d,message one,1,2,3,4,NA two,5,6,,8,world three,9,10,11,12,foo
result = pd.read_csv('ch06/ex5.csv')
result
| something | a | b | c | d | message | |
|---|---|---|---|---|---|---|
| 0 | one | 1 | 2 | 3 | 4 | NaN |
| 1 | two | 5 | 6 | NaN | 8 | world |
| 2 | three | 9 | 10 | 11 | 12 | foo |
pd.isnull(result)
| something | a | b | c | d | message | |
|---|---|---|---|---|---|---|
| 0 | False | False | False | False | False | True |
| 1 | False | False | False | True | False | False |
| 2 | False | False | False | False | False | False |
result = pd.read_csv('ch06/ex5.csv', na_values=['NULL'])
result
| something | a | b | c | d | message | |
|---|---|---|---|---|---|---|
| 0 | one | 1 | 2 | 3 | 4 | NaN |
| 1 | two | 5 | 6 | NaN | 8 | world |
| 2 | three | 9 | 10 | 11 | 12 | foo |
# world를 NA값으로 처리하니 NaN으로 나온다.
# 특정한 값을 NA 처리할 수 있을것 같다.
pd.read_csv('ch06/ex5.csv', na_values=['world'])
| something | a | b | c | d | message | |
|---|---|---|---|---|---|---|
| 0 | one | 1 | 2 | 3 | 4 | NaN |
| 1 | two | 5 | 6 | NaN | 8 | NaN |
| 2 | three | 9 | 10 | 11 | 12 | foo |
열마다 다른 NA 문자를 사전 값으로 넘겨 처리 가능¶
sentinels = {'message': ['foo', 'NA'], 'something': ['two']}
pd.read_csv('ch06/ex5.csv', na_values=sentinels)
| something | a | b | c | d | message | |
|---|---|---|---|---|---|---|
| 0 | one | 1 | 2 | 3 | 4 | NaN |
| 1 | NaN | 5 | 6 | NaN | 8 | world |
| 2 | three | 9 | 10 | 11 | 12 | NaN |
read_csv / read_table 함수 인자¶
| 인자 | 설명 |
|---|---|
| path | 파일 시스템에서의 위치, URL, 파일 객체를 나타내는 문자열 |
| sep or delimiter | 필드를 구분하기 위해 사용할 연속된 문자나 정규표현식 |
| header | 칼럼의 이름으로 사용할 로우의 번호, 기본 값은 0(첫 로우)이며 헤더가 없으면 None으로 지정할 수 있다. |
| index_col | 색인으로 사용할 칼럼 번호나 이름, 계층적 색인을 지정할 경우 리스트를 넘길 수 있다. |
| names | 컬럼 이름으로 사용할 리스트. header = None과 함께 사용한다. |
| skiprows | 파일의 시작부터 무시할 로우의 개수 또는 무시할 로우 번호가 담긴 리스트 |
| na_values | NA 값으로 처리할 값들의 나열 |
| comment | 주석으로 분류되어 파싱하지 않을 문자 혹은 문자열 |
| parse_dates | 날짜를 datetime으로 변환할지의 여부. 기본값은 False이며, True일 경우 모든 칼럼에 다 적용된다. 리스트를 넘기면 변환할 칼럼을 지정할 수 있는데, [1, 2, 3]을 넘기면 각각의 칼럼을 datetime으로 변환하고, [[1, 3]]을 넘기면 1, 3번 칼럼을 조합해서 하나의 datetime으로 변환한다. |
| keep_date_col | 여러 칼럼을 datetime으로 변환했을 경우 원래 칼럼을 남겨둘지의 여부. 기본값은 False |
| converters | 변환 시 칼럼에 적용할 함수를 지정한다. 예를 들어 {'foo': f}는 'foo'칼럼에 f 함수를 적용한다. 전달하는 사전의 키 값은 칼럼 이름이나 번호가 될 수 있다. |
| dayfirst | 모호한 날짜 형식일 경우 국제 형식으로 간주한다(7/6/2012는 2012년 6월 7일로 간주한다). 기본값은 False |
| date_parser | 날짜 변환 시 사용할 함수 |
| nrows | 파일의 첫 일부만 읽어올 때 처음 몇 줄을 읽을 것인지 지정한다. |
| iterator | 파일을 조금씩 읽을 때 사용하도록 TextParser 객체를 반환하도록 한다. 기본값은 False |
| chunksize | TextParser 객체에서 사용할, 한 번에 읽을 파일의 크기 |
| skip_footer | 무시할 파일의 마지막 줄 수 |
| verbose | 파싱 결과에 대한 정보를 출력한다. 숫자가 아닌 값들이 들어있는 칼럼이면서 누락된 값이 있다면 줄 번호를 출력한다. 기본값은 False |
| encoding | 유니코드 인코딩 종류를 지정한다. UTF-8로 인코딩된 텍스트일 경우 'utf-8'로 지정한다. |
| squeeze | 로우가 하나뿐이라면 Series 객체를 반환한다. 기본값은 False |
| thousands | 숫자를 천 단위로 끊을 때 사용할 ', '나 '.' 같은 구분자 |
# 이 명령어로 어떤 함수인지, 어떤 파라미터를 넘겨야 하는지 정확히 알 수 있다.
# 굳이 명령어들을 따라칠 필요는 없는데 어떤 파라미터들을 넘기는지 한 번 공부하는 겸겸해서 쳐봤다.
pd.read_csv?
6.1.1 텍스트 파일 조금씩 읽어오기¶
result = pd.read_csv('ch06/ex6.csv')
result
<class 'pandas.core.frame.DataFrame'> Int64Index: 10000 entries, 0 to 9999 Data columns (total 5 columns): one 10000 non-null values two 10000 non-null values three 10000 non-null values four 10000 non-null values key 10000 non-null values dtypes: float64(4), object(1)
nrows로 처음 몇 줄만 읽어볼 수 있다.¶
- 리눅스의 head 의 기능과 같다고 생각하면 된다.
pd.read_csv('ch06/ex6.csv', nrows=5)
| one | two | three | four | key | |
|---|---|---|---|---|---|
| 0 | 0.467976 | -0.038649 | -0.295344 | -1.824726 | L |
| 1 | -0.358893 | 1.404453 | 0.704965 | -0.200638 | B |
| 2 | -0.501840 | 0.659254 | -0.421691 | -0.057688 | G |
| 3 | 0.204886 | 1.074134 | 1.388361 | -0.982404 | R |
| 4 | 0.354628 | -0.133116 | 0.283763 | -0.837063 | Q |
TextParser 객체를 이용해서 chunksize에 따라 분리된 파일을 순회 가능¶
chunker = pd.read_csv('ch06/ex6.csv', chunksize=1000)
chunker
<pandas.io.parsers.TextFileReader at 0x10c3d2490>
chunker = pd.read_csv('ch06/ex6.csv', chunksize=1000)
tot = Series([])
for piece in chunker:
# piece['key']에 있는 E, X, L 등의 숫자를 센다. 값이 없는 것들은 0으로 채운다.
tot = tot.add( piece['key'].value_counts(), fill_value=0)
# Key가 아닌 값을(order) 기준으로 내림차순 정리
tot = tot.order(ascending=False)
tot[:10]
E 368 X 364 L 346 O 343 Q 340 M 338 J 337 F 335 K 334 H 330 dtype: float64
6.1.2 데이터를 텍스트 형식으로 기록하기¶
data = pd.read_csv('ch06/ex5.csv')
data
| something | a | b | c | d | message | |
|---|---|---|---|---|---|---|
| 0 | one | 1 | 2 | 3 | 4 | NaN |
| 1 | two | 5 | 6 | NaN | 8 | world |
| 2 | three | 9 | 10 | 11 | 12 | foo |
data.to_csv('ch06/out.csv')
!cat ch06/out.csv
,something,a,b,c,d,message 0,one,1,2,3.0,4, 1,two,5,6,,8,world 2,three,9,10,11.0,12,foo
# csv로 지정하는데 output은 표준아웃풋(모니터), separator는 '|'
data.to_csv(sys.stdout, sep='|')
|something|a|b|c|d|message 0|one|1|2|3.0|4| 1|two|5|6||8|world 2|three|9|10|11.0|12|foo
# Write DataFrame to a comma-separated value (csv) file
# na_rep -> Missing data representation. NA REPresentation
data.to_csv?
na_rep로 누락된값을 원하는 값으로 변경 가능¶
data.to_csv(sys.stdout, na_rep='NULL')
,something,a,b,c,d,message 0,one,1,2,3.0,4,NULL 1,two,5,6,NULL,8,world 2,three,9,10,11.0,12,foo
data.to_csv(sys.stdout, na_rep='NaN')
,something,a,b,c,d,message 0,one,1,2,3.0,4,NaN 1,two,5,6,NaN,8,world 2,three,9,10,11.0,12,foo
row, column 값을 저장할 것인지 선택 가능¶
data.to_csv(sys.stdout, index=False, header=False)
one,1,2,3.0,4, two,5,6,,8,world three,9,10,11.0,12,foo
컬럼의 일부분만 기록 가능, 순서를 직접 지정 가능¶
data.to_csv(sys.stdout, index=False, cols=['a', 'b', 'c'])
a,b,c 1,2,3.0 5,6, 9,10,11.0
Series에도 to_csv method 존재¶
dates = pd.date_range('1/1/2000', periods=7)
ts = Series(np.arange(7), index=dates)
ts.to_csv('ch06/tseries.csv')
!cat ch06/tseries.csv
2000-01-01,0 2000-01-02,1 2000-01-03,2 2000-01-04,3 2000-01-05,4 2000-01-06,5 2000-01-07,6
약간 복잡하게 헤더를 없애고 첫 번째 칼럼을 색인으로 하면 read_csv 메서드로 Series 객체를 얻을 수 있지만 from_csv 메서드가 좀 더 편리하고 간단하게 문제 해결¶
pd.DataFrame.to_csv?
Series.from_csv('ch06/tseries.csv', parse_dates=True)
2000-01-01 0 2000-01-02 1 2000-01-03 2 2000-01-04 3 2000-01-05 4 2000-01-06 5 2000-01-07 6 dtype: int64
type( Series.from_csv('ch06/tseries.csv', parse_dates=True) )
pandas.core.series.Series
# parse dates: boolean, default True.
# Parse dates. Different default from read_table
Series.from_csv?
read_csv를 Series로 읽을 수 있다고 실험하는 중인데 잘 안되네..¶
- DataFrame으로 읽어짐
pd.read_csv('ch06/tseries.csv', header=None)
| 0 | 1 | |
|---|---|---|
| 0 | 2000-01-01 | 0 |
| 1 | 2000-01-02 | 1 |
| 2 | 2000-01-03 | 2 |
| 3 | 2000-01-04 | 3 |
| 4 | 2000-01-05 | 4 |
| 5 | 2000-01-06 | 5 |
| 6 | 2000-01-07 | 6 |
type(pd.read_csv('ch06/tseries.csv', header=None))
pandas.core.frame.DataFrame
pd.read_csv?
6.1.3 수동으로 구분 형식 처리하기¶
read_table에서 읽을 수 없는 잘못된 형식의 줄이 포함된 데이터가 드물게 발견 됨 -> 수동 처리¶
!cat ch06/ex7.csv
"a","b","c" "1","2","3" "1","2","3","4"
import csv
f = open('ch06/ex7.csv')
reader = csv.reader(f)
for line in reader:
print line
['a', 'b', 'c'] ['1', '2', '3'] ['1', '2', '3', '4']
lines = list(csv.reader(open('ch06/ex7.csv')))
header, values = lines[0], lines[1:]
header
['a', 'b', 'c']
values
[['1', '2', '3'], ['1', '2', '3', '4']]
# header = a,b,c
# values를 1,1을 같이 묶는다. 2,2 묶고. 3,3 묶고. 4는 header가 a,b,c 3개 밖에 없기 때문에 포함되지 않는다.
data_dict = {h: v for h, v in zip(header, zip(*values))}
data_dict
{'a': ('1', '1'), 'b': ('2', '2'), 'c': ('3', '3')}
CSV 파일은 다양한 파일 존재하기 때문에 다양한 옵션들은 csv.Dialect 상속받아 해결¶
- 다양한 구분자
- 문자열을 둘러싸는 방법
- 개행문자
class my_dialect(csv.Dialect):
lineterminator = '\n'
delimiter = ';'
quotechar = '"'
reader = csv.reader
reader = csv.reader?
reader = csv.reader
reader = csv.reader
TypeError: "quoting" must be an integer¶
# quoting이 꼭 integer여야 한다는 오류가 발생해서 삽질하다가 뒤에 quoting keyword를 붙여줌..
reader = csv.reader(f, dialect=my_dialect)
--------------------------------------------------------------------------- TypeError Traceback (most recent call last) <ipython-input-257-92557f61d368> in <module>() 1 # quoting이 꼭 integer여야 한다는 오류가 발생해서 삽질하다가 뒤에 quoting keyword를 붙여줌.. ----> 2 reader = csv.reader(f, dialect=my_dialect) TypeError: "quoting" must be an integer
reader = csv.reader(f, dialect=my_dialect, quoting=csv.QUOTE_NONE)
csv.QUOTE_NONE
3
reader = csv.reader(f, delimiter='|')
# 어떤 옵션들 있는지 보려고 했더니 안 보여주네...
csv.reader??
CSV Note¶
- 좀 더 복잡하거나 구분자가 한 글자를 초과하는 고정 길이를 가진다면 csv 모듈을 사용할 수 없다.
- 이런 경우에는 줄을 나누고 문자열의 split 메서드나 정규표현식 메서드인 re.split 등을 이용해서 가공하는 작업을 해야 한다.
CSV 파일 기록¶
with open('ch06/mydata.csv', 'w') as f:
writer = csv.writer(f, dialect=my_dialect, quoting=csv.QUOTE_NONE)
writer.writerow(('one', 'two', 'three'))
writer.writerow(('1', '2', '3'))
writer.writerow(('4', '5', '6'))
writer.writerow(('7', '8', '9'))
!cat ch06/mydata.csv
one;two;three 1;2;3 4;5;6 7;8;9
JSON 데이터¶
- JSON(JavaScript Object Notation)은 웹브라우저와 다른 애플리케이션이 HTTP 요청으로 데이터를 보낼 때 널리 사용하는 표준 파일 형식 중 하나다.
- JSON은 CSV 같은 표 형식의 텍스트보다 좀 더 유연한 데이터 형식이며, JSON 데이터의 예는 다음과 같다.
# json은 python에서처럼 '으로 하면 안된다. 현재 """로 감싸 문자열로 저장되어 있기 때문에
# javascript에서는 '를 string 값으로 인식하지 않아서 에러 발생
obj = """
{
'name': 'Wes',
'places_lived': ['United States', 'Spain', 'Germany'],
'pet': null, 'siblings': [{'name': 'Scott', 'age':25, 'pet':'Zuko'},
{'name': 'Katie', 'age':33, 'pet': 'Cisco'}]
}
"""
import json
# ValueError: Expecting property name: line 3 column 5 (char 7)
result = json.loads(obj)
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) <ipython-input-266-f05e1f9794f2> in <module>() 1 # ValueError: Expecting property name: line 3 column 5 (char 7) ----> 2 result = json.loads(obj) /Library/Frameworks/Python.framework/Versions/2.7/lib/python2.7/json/__init__.pyc in loads(s, encoding, cls, object_hook, parse_float, parse_int, parse_constant, object_pairs_hook, **kw) 336 parse_int is None and parse_float is None and 337 parse_constant is None and object_pairs_hook is None and not kw): --> 338 return _default_decoder.decode(s) 339 if cls is None: 340 cls = JSONDecoder /Library/Frameworks/Python.framework/Versions/2.7/lib/python2.7/json/decoder.pyc in decode(self, s, _w) 363 364 """ --> 365 obj, end = self.raw_decode(s, idx=_w(s, 0).end()) 366 end = _w(s, end).end() 367 if end != len(s): /Library/Frameworks/Python.framework/Versions/2.7/lib/python2.7/json/decoder.pyc in raw_decode(self, s, idx) 379 """ 380 try: --> 381 obj, end = self.scan_once(s, idx) 382 except StopIteration: 383 raise ValueError("No JSON object could be decoded") ValueError: Expecting property name: line 3 column 5 (char 7)
obj = """
{
"name": "Wes",
"places_lived": ["United States", "Spain", "Germany"],
"pet": null, "siblings": [{"name": "Scott", "age":25, "pet":"Zuko"},
{"name": "Katie", "age":33, "pet": "Cisco"}]
}
"""
obj
'\n{\n "name": "Wes",\n "places_lived": ["United States", "Spain", "Germany"],\n "pet": null, "siblings": [{"name": "Scott", "age":25, "pet":"Zuko"},\n {"name": "Katie", "age":33, "pet": "Cisco"}]\n}\n'
JSON은 널 값인 null과 다른 몇 가지 사소한 주의사항(리스트의 마지막에 쉼표가 있으면 안된다던가 하는)을 제외하면 파이썬 코드와 거의 유사¶
- 기본 자료형은 객체(사전), 배열(리스트), 문자열, 숫자, 불리언 그리고 널
- 객체의 키는 반드시 문자열
- JSON 읽고 쓸 수 있는 라이브러리가 몇 개 있지만 표준 라이브러리인 json 사용
# ValueError: Expecting property name: line 3 column 5 (char 7)
result = json.loads(obj)
result
{u'name': u'Wes',
u'pet': None,
u'places_lived': [u'United States', u'Spain', u'Germany'],
u'siblings': [{u'age': 25, u'name': u'Scott', u'pet': u'Zuko'},
{u'age': 33, u'name': u'Katie', u'pet': u'Cisco'}]}
json.dumps는 파이썬 객체를 JSON 형태로 변환¶
asjson = json.dumps(result)
# '가 아니라 "인 것을 확인하자
asjson
'{"pet": null, "siblings": [{"pet": "Zuko", "age": 25, "name": "Scott"}, {"pet": "Cisco", "age": 33, "name": "Katie"}], "name": "Wes", "places_lived": ["United States", "Spain", "Germany"]}'
JSON 객체나 객체의 리스트를 DataFrame이나 다른 자료 구조로 어떻게 변환해서 분석을 할 것인지는 독자의 몫¶
- JSON 객체의 리스트를 DataFrame 생성자로 넘기고 데이터 필드 선택 가능
siblings = DataFrame(result['siblings'], columns=['name', 'age'])
siblings
| name | age | |
|---|---|---|
| 0 | Scott | 25 |
| 1 | Katie | 33 |
# 책에 나와있지 않은 내용을 한 번 더 해봐야 쉽게 이해가 되는듯
siblings2 = DataFrame(result['siblings'], columns=['name', 'age', 'pet'])
siblings2
| name | age | pet | |
|---|---|---|---|
| 0 | Scott | 25 | Zuko |
| 1 | Katie | 33 | Cisco |
pandas에서 JSON을 빠르게 읽고(from_json) 쓰는(to_json) 네이티브 구현중¶
6.1.5 XML과 HTML: 웹 내용 긁어오기¶
from lxml.html import parse
from urllib2 import urlopen
# 데이터를 가져 올 url을 넘긴 후
# 데이터를 받아 온 후 parse
parsed = parse(urlopen('http://finance.yahoo.com/q/op?s=AAPL+Options'))
doc = parsed.getroot()
links = doc.findall('.//a')
# 이 객체는 HTML 엘리멘트를 표현하는 객체일뿐
# URL과 링크 이름을 가져오려면 각 엘리먼트에 대해 get 메서드를 호출하여 URL을 얻고
# text_content 메서드를 사용해서 링크 이름을 가져와야 한다.
links[15:20]
[<Element a at 0x10c3e2ad0>, <Element a at 0x10c3e2b30>, <Element a at 0x10c3e2b90>, <Element a at 0x10c3e2bf0>, <Element a at 0x10c3e2c50>]
이 객체는 HTML 엘리먼트를 표현하는 객체일 뿐¶
- 엘리먼트를 표현하는 객체라고 생각하자. 안 그러면 삽질하게 된다!
- URL과 링크 이름을 가져오려면 각 엘리먼트에 대해 get 메서드를 호출하여 URL을 얻고, text_content 메서드를 이용해서 링크 이름을 가져와야 한다.
lnk = links[28]
lnk
<Element a at 0x10c3e2fb0>
lnk.get('href')
'https://edit.yahoo.com/mc2.0/eval_profile?.intl=us&.lang=en-US&.done=http://finance.yahoo.com/q/op%3fs=AAPL%2bOptions&.src=quote&.intl=us&.lang=en-US'
lnk.text_content()
'Account Info'
urls = [lnk.get('href') for lnk in doc.findall('.//a')]
len(urls)
1239
urls[-3:-1]
['http://www.capitaliq.com', 'http://www.csidata.com']
urls[-10:]
['/q?s=AAPL140517P00780000', '/q/op?s=AAPL&k=800.000000', '/q?s=AAPL140517P00800000', '/q/op?s=AAPL&k=805.000000', '/q?s=AAPL140517P00805000', '/q/os?s=AAPL&m=2014-05-30', 'http://help.yahoo.com/l/us/yahoo/finance/quotes/fitadelay.html', 'http://www.capitaliq.com', 'http://www.csidata.com', 'http://www.morningstar.com/']
찾고자 하는 table 일일이 확인¶
- 몇몇 웹사이트는 table마다 id 속성을 줘서 쉽게 할 수 있지만 어디 세상 일이 쉽게 되는게 있나? 노가다 해야지..
tables = doc.findall('.//table')
tables
[<Element table at 0x100769470>, <Element table at 0x1007694d0>, <Element table at 0x100769530>, <Element table at 0x100769590>, <Element table at 0x1007695f0>, <Element table at 0x100769650>, <Element table at 0x1007696b0>, <Element table at 0x100769710>, <Element table at 0x100769770>, <Element table at 0x1007697d0>, <Element table at 0x100769830>, <Element table at 0x100769890>, <Element table at 0x100769ad0>, <Element table at 0x100769b30>, <Element table at 0x100769b90>, <Element table at 0x100769bf0>, <Element table at 0x100769c50>, <Element table at 0x100769cb0>]
calls = tables[9]
calls
<Element table at 0x1007697d0>
puts = tables[13]
rows = calls.findall('.//tr')
rows
[<Element tr at 0x100769d10>, <Element tr at 0x100769d70>, <Element tr at 0x100769e30>, <Element tr at 0x100769e90>, <Element tr at 0x100769ef0>, <Element tr at 0x100769f50>, <Element tr at 0x100769fb0>, <Element tr at 0x100790b90>, <Element tr at 0x100790ad0>, <Element tr at 0x100790050>, <Element tr at 0x1007900b0>, <Element tr at 0x100790110>, <Element tr at 0x100790170>, <Element tr at 0x1007901d0>, <Element tr at 0x100790230>, <Element tr at 0x100790290>, <Element tr at 0x1007902f0>, <Element tr at 0x100790350>, <Element tr at 0x1007903b0>, <Element tr at 0x100790410>, <Element tr at 0x100790470>, <Element tr at 0x1007904d0>, <Element tr at 0x100790530>, <Element tr at 0x100790590>, <Element tr at 0x1007905f0>, <Element tr at 0x100790650>, <Element tr at 0x1007906b0>, <Element tr at 0x100790710>, <Element tr at 0x100790770>, <Element tr at 0x1007907d0>, <Element tr at 0x100790830>, <Element tr at 0x100790890>, <Element tr at 0x1007908f0>, <Element tr at 0x100790950>, <Element tr at 0x1007909b0>, <Element tr at 0x100790a10>, <Element tr at 0x100790a70>, <Element tr at 0x10c3f3d10>, <Element tr at 0x10c3f3d70>, <Element tr at 0x10c3f3dd0>, <Element tr at 0x10c3f3e30>, <Element tr at 0x10c3f3e90>, <Element tr at 0x10c3f3ef0>, <Element tr at 0x10c3f3f50>, <Element tr at 0x10c3f3fb0>, <Element tr at 0x10078d050>, <Element tr at 0x10078d0b0>, <Element tr at 0x10078d110>, <Element tr at 0x10078d170>, <Element tr at 0x10078d1d0>, <Element tr at 0x10078d230>, <Element tr at 0x10078d290>, <Element tr at 0x10078d2f0>, <Element tr at 0x10078d350>, <Element tr at 0x10078d3b0>, <Element tr at 0x10078d410>, <Element tr at 0x10078d470>, <Element tr at 0x10078d4d0>, <Element tr at 0x10078d530>, <Element tr at 0x10078d590>, <Element tr at 0x10078d5f0>, <Element tr at 0x10078d650>, <Element tr at 0x10078d6b0>, <Element tr at 0x10078d710>, <Element tr at 0x10078d770>, <Element tr at 0x10078d7d0>, <Element tr at 0x10078d830>, <Element tr at 0x10078d890>, <Element tr at 0x10078d8f0>, <Element tr at 0x10078d950>, <Element tr at 0x10078d9b0>, <Element tr at 0x10078da10>, <Element tr at 0x10078da70>, <Element tr at 0x10078dad0>, <Element tr at 0x10078db30>, <Element tr at 0x10078db90>, <Element tr at 0x10078dbf0>, <Element tr at 0x10078dc50>, <Element tr at 0x10078dcb0>, <Element tr at 0x10078dd10>, <Element tr at 0x10078dd70>, <Element tr at 0x10078ddd0>, <Element tr at 0x10078de30>, <Element tr at 0x10078de90>, <Element tr at 0x10078def0>, <Element tr at 0x10078df50>, <Element tr at 0x10078dfb0>, <Element tr at 0x100796050>, <Element tr at 0x1007960b0>, <Element tr at 0x100796110>, <Element tr at 0x100796170>, <Element tr at 0x1007961d0>, <Element tr at 0x100796230>, <Element tr at 0x100796290>, <Element tr at 0x1007962f0>, <Element tr at 0x100796350>, <Element tr at 0x1007963b0>, <Element tr at 0x100796410>, <Element tr at 0x100796470>, <Element tr at 0x1007964d0>, <Element tr at 0x100796530>, <Element tr at 0x100796590>, <Element tr at 0x1007965f0>, <Element tr at 0x100796650>, <Element tr at 0x1007966b0>, <Element tr at 0x100796710>, <Element tr at 0x100796770>, <Element tr at 0x1007967d0>, <Element tr at 0x100796830>, <Element tr at 0x100796890>, <Element tr at 0x1007968f0>, <Element tr at 0x100796950>, <Element tr at 0x1007969b0>, <Element tr at 0x100796a10>, <Element tr at 0x100796a70>, <Element tr at 0x100796ad0>, <Element tr at 0x100796b30>, <Element tr at 0x100796b90>, <Element tr at 0x100796bf0>, <Element tr at 0x100796c50>, <Element tr at 0x100796cb0>, <Element tr at 0x100796d10>, <Element tr at 0x100796d70>, <Element tr at 0x100796dd0>, <Element tr at 0x100796e30>, <Element tr at 0x100796e90>, <Element tr at 0x100796ef0>, <Element tr at 0x100796f50>, <Element tr at 0x100796fb0>, <Element tr at 0x100791050>, <Element tr at 0x1007910b0>, <Element tr at 0x100791110>, <Element tr at 0x100791170>, <Element tr at 0x1007911d0>, <Element tr at 0x100791230>, <Element tr at 0x100791290>, <Element tr at 0x1007912f0>, <Element tr at 0x100791350>, <Element tr at 0x1007913b0>, <Element tr at 0x100791410>, <Element tr at 0x100791470>, <Element tr at 0x1007914d0>, <Element tr at 0x100791530>, <Element tr at 0x100791590>, <Element tr at 0x1007915f0>, <Element tr at 0x100791650>, <Element tr at 0x1007916b0>, <Element tr at 0x100791710>, <Element tr at 0x100791770>, <Element tr at 0x1007917d0>, <Element tr at 0x100791830>, <Element tr at 0x100791890>, <Element tr at 0x1007918f0>, <Element tr at 0x100791950>, <Element tr at 0x1007919b0>, <Element tr at 0x100791a10>, <Element tr at 0x100791a70>, <Element tr at 0x100791ad0>, <Element tr at 0x100791b30>, <Element tr at 0x100791b90>, <Element tr at 0x100791bf0>, <Element tr at 0x100791c50>, <Element tr at 0x100791cb0>, <Element tr at 0x100791d10>, <Element tr at 0x100791d70>, <Element tr at 0x100791dd0>, <Element tr at 0x100791e30>, <Element tr at 0x100791e90>, <Element tr at 0x100791ef0>, <Element tr at 0x100791f50>, <Element tr at 0x100791fb0>, <Element tr at 0x100794050>, <Element tr at 0x1007940b0>, <Element tr at 0x100794110>, <Element tr at 0x100794170>, <Element tr at 0x1007941d0>, <Element tr at 0x100794230>, <Element tr at 0x100794290>, <Element tr at 0x1007942f0>, <Element tr at 0x100794350>, <Element tr at 0x1007943b0>, <Element tr at 0x100794410>, <Element tr at 0x100794470>, <Element tr at 0x1007944d0>, <Element tr at 0x100794530>, <Element tr at 0x100794590>, <Element tr at 0x1007945f0>, <Element tr at 0x100794650>, <Element tr at 0x1007946b0>, <Element tr at 0x100794710>, <Element tr at 0x100794770>, <Element tr at 0x1007947d0>, <Element tr at 0x100794830>, <Element tr at 0x100794890>, <Element tr at 0x1007948f0>, <Element tr at 0x100794950>, <Element tr at 0x1007949b0>, <Element tr at 0x100794a10>, <Element tr at 0x100794a70>, <Element tr at 0x100794ad0>, <Element tr at 0x100794b30>, <Element tr at 0x100794b90>, <Element tr at 0x100794bf0>, <Element tr at 0x100794c50>, <Element tr at 0x100794cb0>, <Element tr at 0x100794d10>, <Element tr at 0x100794d70>, <Element tr at 0x100794dd0>, <Element tr at 0x100794e30>, <Element tr at 0x100794e90>, <Element tr at 0x100794ef0>, <Element tr at 0x100794f50>, <Element tr at 0x100794fb0>, <Element tr at 0x100797050>, <Element tr at 0x1007970b0>, <Element tr at 0x100797110>, <Element tr at 0x100797170>, <Element tr at 0x1007971d0>, <Element tr at 0x100797230>, <Element tr at 0x100797290>, <Element tr at 0x1007972f0>, <Element tr at 0x100797350>, <Element tr at 0x1007973b0>, <Element tr at 0x100797410>, <Element tr at 0x100797470>, <Element tr at 0x1007974d0>, <Element tr at 0x100797530>, <Element tr at 0x100797590>, <Element tr at 0x1007975f0>, <Element tr at 0x100797650>, <Element tr at 0x1007976b0>, <Element tr at 0x100797710>, <Element tr at 0x100797770>, <Element tr at 0x1007977d0>, <Element tr at 0x100797830>, <Element tr at 0x100797890>, <Element tr at 0x1007978f0>, <Element tr at 0x100797950>, <Element tr at 0x1007979b0>, <Element tr at 0x100797a10>, <Element tr at 0x100797a70>, <Element tr at 0x100797ad0>]
웹페이지 구조가 안 바뀌었네.¶
- 책을 쓴게 2012년 10월 29일인데 아직까지 안 바뀌다니...
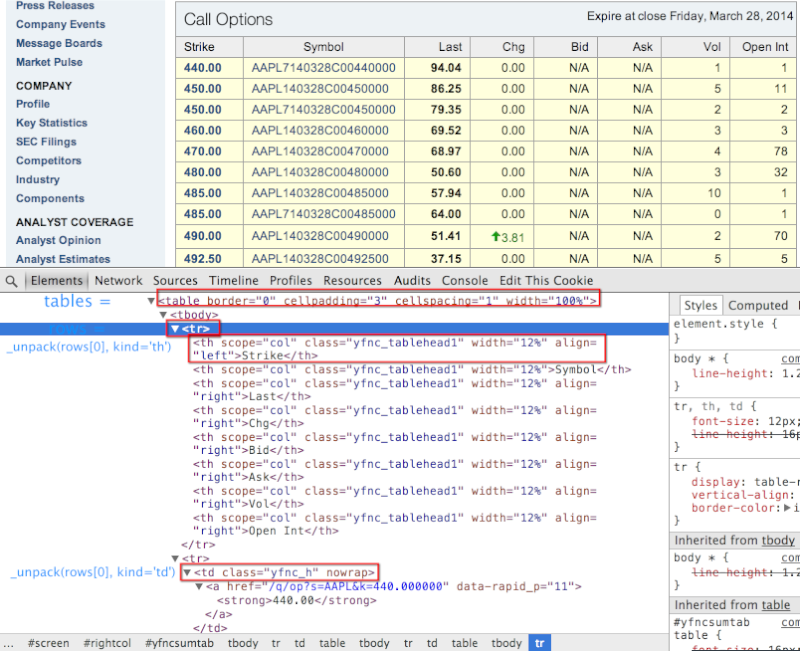
def _unpack(row, kind='td'):
elts = row.findall('.//%s' % kind)
return [val.text_content() for val in elts]
_unpack(rows[0], kind='th')
['Strike', 'Symbol', 'Last', 'Chg', 'Bid', 'Ask', 'Vol', 'Open Int']
_unpack(rows[1], kind='td')
['330.00', 'AAPL140517C00330000', '263.00', ' 0.00', '255.70', '258.25', '6', '2']
Yahoo finance HTML structure in Chrome Developer Tools¶
- 처음 보는 사람은 잘 이해가 안 될 것이다.
- Yahoo 사이트의 HTML 구조 먼저 파악을 하고 시작하자.
- 이 구조가 이해 안되면 2번 봐라.
단계들을 통합하여 웹에서 긁어온 데이터를 DataFrame으로 변환¶
- 숫자 데이터지만 여전히 문자열 형식으로 저장되어 있으므로 적절하게 변환을 해줘야 하는데 모든 데이터가 실수형은 아닐 것이므로 이 작업은 수동으로 처리
- 하지만 운 좋겠도 pandas에는 TextParser 클래스가 있어 자동 형 변환을 적절하게 수행해준다.
- TextParser 클래스는 read_csv 함수와 다른 파싱 함수에서도 사용
from pandas.io.parsers import TextParser
def parse_options_data(table):
rows = table.findall('.//tr')
# rows[0]은 header
header = _unpack(rows[0], kind='th')
# rows[1:] 부터 실제적인 data
data = [_unpack(r) for r in rows[1:]]
# TextParser에 data를 넘기고 column명으로 header를 사용
return TextParser(data, names=header).get_chunk()
마지막으로 lxml 테이블 객체를 위에서 작성한 파싱 함수를 이용해서 처리하면 DataFrame 결과값 얻을 수 있다¶
[옵션(금융) - wiki kr](http://ko.wikipedia.org/wiki/%EC%98%B5%EC%85%98_(%EA%B8%88%EC%9C%B5)¶
- 금융 데이터를 분석하는 것이기 때문에 금융 파트에 대한 도메인 지식이 있어야 한다. 내가 분석하려는 데이터가 어떠한 역할을 하는지 모르면 말짱 황!
- 옵션(option)은 파생 상품의 일종이며, 미리 결정된 기간 안에 특정 상품을 정해진 가격으로 사고 팔 수 있는 권리를 말한다.
- call option: 특정 금융 상품을 정해진 가격에 매입할 수 있는 권리를 가진 매입 옵션(call option). 시장에서 내가 얼마 줄테니 팔아라 하는 형식이라 call 이라 부르는듯
- put option: 매도할 수 있는 권리를 가진 매도 옵션(put option)으로 나뉜다. put. 밀다. 시장에 내가 얼마에 팔겠다고 민다. 라고 생각하면 편할듯
# call option data
call_data = parse_options_data(calls)
# put option data
put_data = parse_options_data(puts)
call_data[:10]
| Strike | Symbol | Last | Chg | Bid | Ask | Vol | Open Int | |
|---|---|---|---|---|---|---|---|---|
| 0 | 330 | AAPL140517C00330000 | 263.00 | 0.00 | 255.70 | 258.25 | 6 | 2 |
| 1 | 400 | AAPL140517C00400000 | 193.00 | 0.00 | 186.40 | 187.95 | 1 | 9 |
| 2 | 410 | AAPL140517C00410000 | 181.30 | 0.00 | 175.70 | 178.10 | 68 | 10 |
| 3 | 420 | AAPL140517C00420000 | 170.80 | 0.00 | 165.85 | 168.65 | 124 | 1 |
| 4 | 430 | AAPL140517C00430000 | 160.75 | 0.00 | 155.85 | 158.05 | 376 | 2 |
| 5 | 440 | AAPL140517C00440000 | 152.87 | 0.00 | 146.35 | 147.90 | 1 | 1 |
| 6 | 445 | AAPL140517C00445000 | 145.55 | 0.00 | 140.95 | 143.25 | 106 | 1 |
| 7 | 450 | AAPL140517C00450000 | 137.00 | 5.00 | 136.35 | 137.90 | 1 | 48 |
| 8 | 450 | AAPL7140517C00450000 | 137.86 | 2.39 | 135.05 | 138.80 | 1 | 1 |
| 9 | 455 | AAPL140517C00455000 | 138.00 | 0.00 | 131.35 | 132.90 | 2 | 2 |
put_data[:10]
| Strike | Symbol | Last | Chg | Bid | Ask | Vol | Open Int | |
|---|---|---|---|---|---|---|---|---|
| 0 | 280 | AAPL140517P00280000 | 0.05 | 0 | N/A | 0.04 | 2 | 6 |
| 1 | 290 | AAPL140517P00290000 | 0.02 | 0 | N/A | 0.01 | 11 | 11 |
| 2 | 295 | AAPL140517P00295000 | 0.01 | 0 | N/A | 0.04 | 3 | 8 |
| 3 | 300 | AAPL140517P00300000 | 0.05 | 0 | N/A | 0.01 | 1 | 23 |
| 4 | 305 | AAPL140517P00305000 | 0.05 | 0 | N/A | 0.01 | 10 | 20 |
| 5 | 310 | AAPL140517P00310000 | 0.10 | 0 | N/A | 0.04 | 0 | 1 |
| 6 | 315 | AAPL140517P00315000 | 0.12 | 0 | N/A | 0.04 | 0 | 1 |
| 7 | 320 | AAPL140517P00320000 | 0.02 | 0 | N/A | 0.04 | 1 | 7 |
| 8 | 325 | AAPL140517P00325000 | 0.05 | 0 | N/A | 0.01 | 185 | 342 |
| 9 | 330 | AAPL140517P00330000 | 0.02 | 0 | N/A | 0.04 | 5 | 5 |
lxml.objectify 이용해 XML 파싱하기¶
- XML(eXtensible Markup Language)은 계층적 구조와 메타데이터를 포함하는 중첩된 데이터 구조를 지원하는 또 다른 유명한 데이터 형식이다. 지금 이 책도 실제로는 XML 문서로 작성
- 뉴욕 MTA(Metropolitan Transportation Authority)는 버스와 전철 운영에 관한 여러 가지 데이터 공개
- 살펴볼 것은 여러 XML 파일로 제공되는 실적 자료
- 전철과 버스 운영은 매월 아래와 비슷한 내용의 각각 다른 파일(Metro-North Railroad의 경우 Preformance_MNR.xml 같은)로 제공
- Preformance_MNR.xml 이 파일을 못 찾았는데 임주영님께서 찾아서 올려주셨습니다. github ch06 디렉토리에 저장해 두었습니다. 그런데 계속 오류가 나는군요. < PERFORMANCE > 이 태그 때문인듯한데. 해결책 알고 계신분은 좀 알려주세요. 계속 오류가 발생하여 책에 나와있는 것으로 테스트 하였습니다.
Performance_MNR.xml을 어떻게 받는지 모르겠다.¶
- 소스파일을 6장을 뒤져봐도 없고
- 홈페이지에는 아마 XML 구조가 바뀐듯 싶다.
- 그래서 최후의 수단으로 얼마 안되서 그냥 내가 일일이 쳤다.
- XML은 엄격하기 때문에 하나라도 오타가 있으면 오류 발생하므로 주의!
%%writefile ch06/Performance_MNR.xml
<INDICATOR>
<INDICATOR_SEQ>373889</INDICATOR_SEQ>
<PARENT_SEQ></PARENT_SEQ>
<AGENCY_NAME>MEtro-North Railroad</AGENCY_NAME>
<INDICATOR_NAME>Escalator Availability</INDICATOR_NAME>
<DESCRIPTION>Percent of the time that escalators are operational systemwide. The availability rate is based on physical observations performed the morning of regular business days only. This is a new indicator the agency began reporting in 2009.</DESCRIPTION>
<PERIOD_YEAR>2011</PERIOD_YEAR>
<PERIOD_MONTH>12</PERIOD_MONTH>
<CATEGORY>Service Indicators</CATEGORY>
<FREQUENCY>M</FREQUENCY>
<DESIRED_CHANGE>U</DESIRED_CHANGE>
<INDICATOR_UNIT>%</INDICATOR_UNIT>
<DECIMAL_PLACES>1</DECIMAL_PLACES>
<YTD_TARGET>97.00</YTD_TARGET>
<YTD_ACTUAL></YTD_ACTUAL>
<MONTHLY_TARGET>97.00</MONTHLY_TARGET>
<MONTHLY_ACTUAL></MONTHLY_ACTUAL>
</INDICATOR>
Overwriting ch06/Performance_MNR.xml
from lxml import objectify
import urllib2
path = 'Performance_MNR.xml'
# online_path = 'http://www.mta.info/developers/data/lirr/lirr_gtfs.xml'
# data = urllib2.urlopen(online_path).read()
# f = open(path, 'w')
# f.write(data)
# f.close()
# objectify를 이용해서 파일 파싱
parsed = objectify.parse(open(path))
root = parsed.getroot()
--------------------------------------------------------------------------- IOError Traceback (most recent call last) <ipython-input-304-a212823090af> in <module>() 11 12 # objectify를 이용해서 파일 파싱 ---> 13 parsed = objectify.parse(open(path)) 14 root = parsed.getroot() IOError: [Errno 2] No such file or directory: 'Performance_MNR.xml'
data = []
skip_fields = ['PARENT_SEQ', 'INDICATOR_SEQ',
'DESIRED_CHANGE', 'DECIMAL_PLACES']
root.INDICATOR를 통해 모든 엘리먼트를 끄집어 낼 수 있다
- 각각의 항목에 대해 몇몇 태그는 제외하고 태그 이름(YTD_ACTUAL 같은)을 키 값으로 하는 사전을 만들어 냄
# root.INDICATOR -> root
for elt in root:
el_data = {}
for child in elt.getchildren():
if child.tag in skip_fields:
continue
el_data[child.tag] = child.pyval
data.append(el_data)
data
# 위의 값과 비교하기 위해 테스트 해본 것
for elt in root:
for child in elt.getchildren():
print child.tag, child.pyval
5장에서 사전 형식은 DataFrame으로 변환할 수 있다는 것 참고¶
perf = DataFrame(data)
perf
6.2 이진 데이터 형식¶
데이터를 효율적으로 저장하는 가장 손쉬운 방법¶
- 파이썬에 기본으로 내장되어 있는 pickle 직렬화를 통해 데이터를 이진 형식으로 저장하는 것
- 편리하게도 pandas의 객체는 모두 pickle을 이용해서 데이터를 저장하는 save 메서드 있음
frame = pd.read_csv('ch06/ex1.csv')
frame
frame.save('ch06/frame_pickle')
pd.load('ch06/frame_pickle')
pickle 사용시 주의사항¶
- pickle은 오래 보관할 필요가 없는 데이터에만 추천
- 오랜 시간이 지나도 안정적으로 데이터를 저장할 거라고 보장 못함
6.2.1 HDF5 형식 사용하기¶
디스크에 이진 형식으로 저장된 대용량의 과학 자료를 효율적으로 읽고 쓸 수 있는 다양한 도구 존재
산업 기준에 맞는 인기 라이브러리중 하나가 HDF5(Hierarchical Data Format), 계층적 데이터 형식
내부적으로 파일 시스템 같은 노드 구조
여러 개의 데이터셋을 저장하고 부가 정보 기록 가능
다양한 압축 기술을 사용해서 on-the-fly(실시간) 압축 지원
반복되는 패턴을 가진 데이터 좀 더 효과적 저장
메모리에 모두 적재할 수 없는 엄청나게 큰 데이터를 아주 큰 배열에서 필요한 만큼의 작은 부분들만 효과적으로 읽고 쓸 수 있는 훌륭한 선택
PyTables: HDF5를 추상화하여 여러가지 유연한 데이터 컨테이너와 테이블 색인, 질의 기능 그리고 외부 메모리 연산(out-of-core, external memory algorithm) 지원
h5py: 직접적이지만 고수준의 HDF5 API에 대한 인터페이스 제공
pandas는 PyTable를 이용한 HDFStore라는 가벼운 사전 클래스를 통해 pandas 객체를 저장
PyTables를 설치 안했을 때 나오는 상황¶
그럼 일반적인 사용자들은 numexpr 인가? 이 라이브러리가 또 없다고 한다. 그럼 이 라이브러리를 먼저 설치하고 tables를 설치한다.
설치 방법1: sudo easy_install tables(오류 확인)
설치 방법2: sudo easy_install numexpr(확실하지 않음. 확인 요함)
설치 방법3: sudo easy_install tables
---------------------------------------------------------------------------
Exception Traceback (most recent call last)
<ipython-input-135-35f4287dfd8f> in <module>()
----> 1 store = pd.HDFStore('ch06/mydata.h5')
/Library/Python/2.7/site-packages/pandas-0.12.0_307_g3a2fe0b-py2.7-macosx-10.8-intel.egg/pandas/io/pytables.pyc in __init__(self, path, mode, complevel, complib, fletcher32, **kwargs)
343 import tables as _
344 except ImportError: # pragma: no cover
--> 345 raise Exception('HDFStore requires PyTables')
346
347 self._path = path
Exception: HDFStore requires PyTables
HDFStore를 사용하기 위해서는 PyTables 라이브러리를 설치해야 한다.¶
- 이런게 있다는 것을 indexing 해두고 나중에 필요하면 찾아보자!
# 라이브러리 설치해보고 테스트 해보라.
store = pd.HDFStore('ch06/mydata.h5')
store['obj1'] = frame
store['obj1_col'] = frame['a']
store
store['obj1']
데이터 분석 문제¶
- 대부분 CPU보다는 IO 성능에 의존적
- HDF5는 데이터베이스가 아니다. HDF5는 한 번만 기록하고 여러 번 자주 읽어야 하는 데이터에 최적화되어 있다. 데이터는 아무때나 파일에 추가할 수 있지만 만약 여러 곳에서 동시에 파일을 쓴다면 파일이 깨지는 문제가 발생할 수 있다.
6.2.2 마이크로소프트 엑셀 파일에서 데이터 읽어오기¶
- pandas는 ExcelFile 클래스를 통해 마이크로소프트 엑셀 2003 이후 버전의 데이터를 읽기 가능
- 내부적으로 ExcelFile 클래스는 xlrd, openpyxl 패키지 활용. 사용하기 전에 먼저 설치
Excel 작업시 주의사항¶
- 현업에서는 Excel 에서 오류가 많이 발생하기 때문에 csv로 변경 후에 작업한다고 한다. 그러니 굳이 excel 파일로 하여 Error를 만들지 말고 안전하게 csv 파일로 변경 후에 사용하자.
- 이런것이 있다 정도만 알아놓자.
- 여기는 그냥 skip 하겠다.
xls_file = pd.ExcelFile('data.xls')
table = xls_file.parse('Sheet1')
6.3 HTML, 웹 API와 함꼐 사용하기¶
Requests: HTTP for Humans¶
- urllib2보다 더 간편
- similar code, without Requests.
트위터 분석 문제 발생¶
- 트위터는 처음에 아무런 인증없이 API를 제공하다 망 과부하가 발생하자 OAuth 인증 방식으로 변경
- OAuth2 - API 인증을 위한 만능도구상자
- Twitter API
- 지금은 인증 문제 때문에 pass 하겠음
import requests
url = 'http://search.twitter.com/search.json?q=python%20pandas'
resp = requests.get(url)
resp
<Response [401]>
resp.text
u'{"errors":[{"message":"The Twitter REST API v1 is no longer active. Please migrate to API v1.1. https://dev.twitter.com/docs/api/1.1/overview.","code":64}]}'
조금만 수고를 하면 평범한 웹 API를 위한 고수준의 인터페이스를 만들어서 DataFrame에 저장하고 쉽게 분석 작업 수행 가능¶
6.4 데이터베이스와 함께 사용하기¶
- 대부분의 애플리케이션은 텍스트 파일에서 데이터를 읽어오지 않음
- 왜냐하면 대용량의 데이터를 저장하기에 텍스트 파일은 상당히 비효율적
- SQL 기반의 관계형 데이터 베이스가 많이 사용됨. MySql 같은
- 최근 유명해진 NoSQL이라 불리는 비 SQL 기반의 데이터베이스도 많이 사용됨
- SQL vs NoSQL은 서로 각각의 장점을 파악하고 자신의 업무에 맞는 DB를 선택
- SQL에서 데이터를 읽어와서 DataFrame에 저장하는 방법은 꽤 직관적
import sqlite3
query = """
CREATE TABLE test
(a VARCHAR(20), b VARCHAR(20),
c REAL, d INTEGER
);"""
con = sqlite3.connect(':memory:')
con.execute(query)
con.commit()
data = [('Atlanta', 'Georgia', 1.25, 6),
('Tallahassee', 'Florida', 2.6, 3),
('Sacramento', 'California', 1.7, 5)]
stmt = "INSERT INTO test VALUES(?, ?, ?, ?)"
con.executemany(stmt, data)
con.commit()
대부분의 파이썬 SQL 드라이버(PyODBC, psycopg2, MySQLdb, pymssql 등)는 테이블에 대해 select 쿼리를 수행하면 튜플 리스트를 반환한다
cursor = con.execute('select * from test')
rows = cursor.fetchall()
rows
[(u'Atlanta', u'Georgia', 1.25, 6), (u'Tallahassee', u'Florida', 2.6, 3), (u'Sacramento', u'California', 1.7, 5)]
반환된 튜플 리스트를 DataFrame 생성자에 바로 전달해도 되지만 칼럼의 이름을 지정해주면 더 편하다. cursor의 description 속성을 활용하자.
This read-only attribute is a list of 7-item tuples, each containing (name, type_code, display_size, internal_size, precision, scale, null_ok). pyodbc only provides values for name, type_code, internal_size, and null_ok. The other values are set to None.
This attribute will be None for operations that do not return rows or if one of the execute methods has not been called.
The type_code member is the class type used to create the Python objects when reading rows. For example, a varchar column's type will be str.
cursor.desccription 이 아직도 뭔지 잘 모르겠다. 왜 a, b, c, d로 정해진 것도 잘 모르겠고..¶
cursor.description?
Type: tuple
String form: (('a', None, None, None, None, None, None), ('b', None, None, None, None, None, None), ('c', None, None, None, None, None, None), ('d', None, None, None, None, None, None))
Length: 4
Docstring:
tuple() -> empty tuple
tuple(iterable) -> tuple initialized from iterable's items
If the argument is a tuple, the return value is the same object.
cursor.description
(('a', None, None, None, None, None, None),
('b', None, None, None, None, None, None),
('c', None, None, None, None, None, None),
('d', None, None, None, None, None, None))
# cursor.description을 여러개 받아서 0번째 값들을 zip으로 묶는다.
zip(*cursor.description)[0]
('a', 'b', 'c', 'd')
DataFrame(rows, columns=zip(*cursor.description)[0])
| a | b | c | d | |
|---|---|---|---|---|
| 0 | Atlanta | Georgia | 1.25 | 6 |
| 1 | Tallahassee | Florida | 2.60 | 3 |
| 2 | Sacramento | California | 1.70 | 5 |
DataFrame(rows, columns=zip(*cursor.description)[1])
| None | None | None | None | |
|---|---|---|---|---|
| 0 | Atlanta | Georgia | 1.25 | 6 |
| 1 | Tallahassee | Florida | 2.60 | 3 |
| 2 | Sacramento | California | 1.70 | 5 |
# readonly attribute 란다.
# 난 cursor.description을 수정해서 내가 원하는 컬럼값으로 변경하려고 했는데..
# 그럼 어떻게 변경을 해야하지?
cursor.description = '1'
--------------------------------------------------------------------------- TypeError Traceback (most recent call last) <ipython-input-317-4bf5306f81d9> in <module>() 2 # 난 cursor.description을 수정해서 내가 원하는 컬럼값으로 변경하려고 했는데.. 3 # 그럼 어떻게 변경을 해야하지? ----> 4 cursor.description = '1' TypeError: readonly attribute
# 그냥 column에 내가 쓰고 싶은것 정하면 되네..
DataFrame(rows, columns=['country', 'state', 'grade1', 'grade2'])
| country | state | grade1 | grade2 | |
|---|---|---|---|---|
| 0 | Atlanta | Georgia | 1.25 | 6 |
| 1 | Tallahassee | Florida | 2.60 | 3 |
| 2 | Sacramento | California | 1.70 | 5 |
- 데이터베이스에 쿼리를 보내려고 매번 이렇게 하는건 너무 귀찮음
- pandas.io.sql 모듈의 read_frame 함수를 이용하면 간편하게 해결
- 그냥 select 쿼리문과 데이터 베이스 연결 객체(con)만 넘기면 된다
import pandas.io.sql as sql
sql.read_frame('select * from test', con)
| a | b | c | d | |
|---|---|---|---|---|
| 0 | Atlanta | Georgia | 1.25 | 6 |
| 1 | Tallahassee | Florida | 2.60 | 3 |
| 2 | Sacramento | California | 1.70 | 5 |
6.4.1 MongoDB에 데이터 저장하고 불러오기¶
- NoSQL 데이터베이스는 매우 다양한 형태
- 버클리DB나 도쿄캐비닛 같은 것은 사전처럼 키-값을 저장하기도 하고
- 또 다른 것은 기본 저장소는 사전 같은 객체를 사용하며 문서 기반으로 데이터를 저장하기도 한다.
- 이 책에서는 MongoDB를 예제로 선택
- MongoDB 서버를 로컬에 설치하고 공식 드라이버인 pymongo를 사용해서 기본 포트로 번호로 연결
- 현재 필자의 컴퓨터에는 아직 설치하지 않음. 이런 형식으로 한다는 느낌만 가지자
스터디할 때 나왔던 주의사항들¶
- 현업에서는 read_csv나 read_table을 많이 쓴다.
- 엑셀 -> csv로 변환 후 사용한다.
- 왜냐하면 csv에 훨씬 강력한 라이브러리들이 있는데 굳이 엑셀을 활용할 필요가 없다. 또한 엑셀은 보이지 않는 값들이 많아 오류의 주범
- sep에 Regular Expression을 사용할 수 있다.
- 엑셀에서 XML 데이터 가져오기