CS579: Lecture 05¶
** Community Detection **
Dr. Aron Culotta
Illinois Institute of Technology
(Many figures come from Mining of Massive Datasets, Jure Leskovec, Anand Rajaraman, Jeff Ullman)

- Why do we want to identify communities?
- What are the "communities" in this graph?
- Why did you choose these communities?
A bad solution: Agglomerative clustering
- Let distance function d(A,B) be the shortest path between nodes A and B
- Let Ci and Cj be two clusters of nodes. Then, let the distance between two clusters be the minimum distance of any two nodes: d(Ci,Cj)=minX∈Ci,Y∈Cjd(X,Y)
- Greedy agglomerative clustering iterative merges the closest two clusters
What would agglomerative clustering do on this network?

d(A,B)=d(A,C)=d(B,C)=d(B,D)=d(D,E)=d(D,F)=d(D,G)=d(E,F)=d(G,F)=1
d(A,D)=d(C,D)...=2
First merge: sample randomly from all nodes with distance == 1.
So, 19 chance we merge B and D in first merge.
Not desireable...any other ideas?
What makes the edge between B and D special?
Betweenness: The betweenness of an edge (A,B) is the number of shortest paths between any nodes X and Y that include edge (A,B).
High betweenness → A and B belong in different communities.

What is betweenness of (B,D)?
(B,D) is on all shortest paths connecting any of {A,B,C} to any of {D,E,F,G}.
Thus, total number of shortest paths = number passing through (B,D) = 3∗4=12.. So, bt(B,D)=12
What is betweenness of (D,F)?
(D,F) is on shortest paths from {A,B,C,D} to {F}.
Thus, betweenness is 4∗1=4.
Consider this graph:

What is betweenness of (D,G)?
(D,G) is on the shortest path from D to G.
(D,G) is also on one of the two shortest paths from D to F and E to G.
Since there can be several shortest paths between two nodes, an edge (a, b) is credited with the fraction of those shortest paths that include the edge (a, b).
Thus, betweenness is 12+12+1=2.
Formally:
bt(e)=∑s,t∈Vσ(s,t|e)σ(s,t)where
- V is the set of nodes
- σ(s,t) is the number of shortest paths between nodes s and t
- σ(s,t|e) is the number of those paths passing through some edge e
If s=t, σ(s,t)=1
So, if there are two shortest paths between s and t, but only one goes through e, betweenness only increases by 0.5.
import networkx as nx
%matplotlib inline
def create_example_graph():
graph = nx.Graph()
graph.add_edges_from([('A', 'B'), ('A', 'C'), ('B', 'C'),
('B', 'D'), ('D', 'E'), ('D', 'F'),
('D', 'G'), ('E', 'F'), ('G', 'F')])
return graph
graph = create_example_graph()
nx.draw(graph, with_labels=True)
# We'll use networkx's built-in betweenness computation in this example.
nx.edge_betweenness_centrality(graph, normalized=False)
# nx.edge_betweenness_centrality(graph, normalized=True)
# normalized between 0-1
{('A', 'B'): 5.0,
('A', 'C'): 1.0,
('B', 'C'): 5.0,
('B', 'D'): 12.0,
('D', 'E'): 4.5,
('D', 'F'): 4.0,
('D', 'G'): 4.5,
('E', 'F'): 1.5,
('F', 'G'): 1.5}
** How to compute shortest path in undirected graph? **
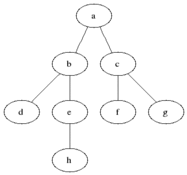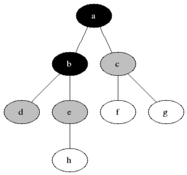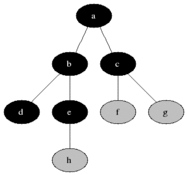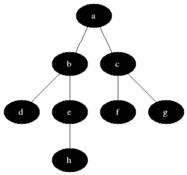
** Breadth first search: ** Given a sourse node s, compute the shortest paths to each of its neighbors. Proceed
from collections import deque
# double ended queue
# stored as a doubly linked list
q = deque()
q.append(1)
print(q)
q.append(2)
print(q)
q.append(3)
print(q)
print('popleft returns: %d' % q.popleft())
print(q)
print('pop returns: %d' % q.pop())
print(q)
deque([1]) deque([1, 2]) deque([1, 2, 3]) popleft returns: 1 deque([2, 3]) pop returns: 3 deque([2])
# compare with:
a = [1,2,3]
print(a.pop(0))
#print(a.pop())
a
1
[2, 3]
What is running time to remove first element of a dynamic array with n elements (a list in Python)?
O(n): Need to shift all elements to the left.
What is the running time to remove first element of a doubly linked list n elements (a deque in Python)?
O(1)
# Sample implementation of doubly linked list.
class Node:
def __init__(self, val):
self.val = val
self.prev = None # previous node
self.next = None # next node
self.head = self
def display(self):
node = self.head
vals = []
while node:
vals.append(node.val)
node = node.next
print(vals)
def popleft(self):
"""
Remove leftmost element of list.
"""
v = self.head.val
self.next.prev = None
self.head = self.next
return v
n1 = Node(1)
n2 = Node(2)
n3 = Node(3)
n1.next = n2
n2.prev = n1
n2.next = n3
n3.prev = n2
mylist = n1
mylist.display()
print(mylist.popleft())
print(mylist.popleft())
[1, 2, 3] 1 2
# to get the neighbors of a node:
list(graph.neighbors('A'))
# vs
#graph.neighbors('A')
['B', 'C']
nx.draw(graph, with_labels=True)
def bfs(graph, start):
"""
Return the order in which nodes are visited in a breadth-first
traversal, starting with the given node.
"""
q = deque()
q.append(start)
seen = set() # nodes we have already visited.
res = []
while len(q) > 0: # while more to visit
n = q.popleft()
if n not in seen:
res.append(n)
seen.add(n)
for nn in graph.neighbors(n):
if nn not in seen:
q.append(nn)
return res
bfs(graph, 'D')
['D', 'B', 'E', 'F', 'G', 'A', 'C']
To get all shortest paths from a node, perform BFS, while keeping track of the depth of the search.
for s in graph.nodes():
paths = nx.single_source_shortest_path(graph, s)
print('\nshortest paths for %s' % s)
print(paths)
shortest paths for A
{'A': ['A'], 'B': ['A', 'B'], 'C': ['A', 'C'], 'D': ['A', 'B', 'D'], 'E': ['A', 'B', 'D', 'E'], 'F': ['A', 'B', 'D', 'F'], 'G': ['A', 'B', 'D', 'G']}
shortest paths for B
{'B': ['B'], 'A': ['B', 'A'], 'C': ['B', 'C'], 'D': ['B', 'D'], 'E': ['B', 'D', 'E'], 'F': ['B', 'D', 'F'], 'G': ['B', 'D', 'G']}
shortest paths for C
{'C': ['C'], 'A': ['C', 'A'], 'B': ['C', 'B'], 'D': ['C', 'B', 'D'], 'E': ['C', 'B', 'D', 'E'], 'F': ['C', 'B', 'D', 'F'], 'G': ['C', 'B', 'D', 'G']}
shortest paths for D
{'D': ['D'], 'B': ['D', 'B'], 'E': ['D', 'E'], 'F': ['D', 'F'], 'G': ['D', 'G'], 'A': ['D', 'B', 'A'], 'C': ['D', 'B', 'C']}
shortest paths for E
{'E': ['E'], 'D': ['E', 'D'], 'F': ['E', 'F'], 'B': ['E', 'D', 'B'], 'G': ['E', 'D', 'G'], 'A': ['E', 'D', 'B', 'A'], 'C': ['E', 'D', 'B', 'C']}
shortest paths for F
{'F': ['F'], 'D': ['F', 'D'], 'E': ['F', 'E'], 'G': ['F', 'G'], 'B': ['F', 'D', 'B'], 'A': ['F', 'D', 'B', 'A'], 'C': ['F', 'D', 'B', 'C']}
shortest paths for G
{'G': ['G'], 'D': ['G', 'D'], 'F': ['G', 'F'], 'B': ['G', 'D', 'B'], 'E': ['G', 'D', 'E'], 'A': ['G', 'D', 'B', 'A'], 'C': ['G', 'D', 'B', 'C']}
Girvan-Newman Algorithm¶
Input: Graph G; desired number of clusters k
Output: A hierarchical clustering of nodes, based on edge betweenness
- While number of clusters <k:
- Compute the betweenness of all edges in G
- Remove edge with highest betweenness


Computing betweenness of all edges¶
- All pairs-shortest-paths, but need to store the paths.
- How can we reduce redundant computation?
Computing betweenness of all edges¶

1.) Do breadth-first search starting at node E.
- Each level is length of shortest path from E to that node
- Edges within the same level cannot be part of a shortest path from E to some target.
2.) Label each node by the number of shortest paths that reach it from the root.
- Start by labeling the root (E). Then, each child node is the sum of its parents.
- E.g., G=D+F
Computing betweenness of all edges¶

3.) Compute fraction of shortest paths through each edge (bottom up).
- leaf nodes get credit 1
- non-leaf nodes get credit of 1 + credits for edges to nodes at level below
- edges to level above gets credit proportional to fraction of shortest paths that go through it.
E.g. Level 3:
- A and C are given credit 1 (they are leaf nodes)
Level 2:
- B gets credit 3 (A+C+1)
- All shortest paths from {E} to {A,B,C} go through B.
- G gets credit 1 (leaf)
Computing betweenness of all edges¶

Level 1 Edges:
- D,B edge gets all credit from node B (3)
- G has two parents, so edges (D,G), (F,G) share the credit from G
- From step 1, D and F each have credit 1, so shared equally. (11+1=.5)
- What if D=5, F=3? 58, 38
Level 1 Nodes:
- D=1+3+.5=4.5
- F=1+.5=1.5
Final steps:
- Repeat for each node as source
- Divide total by 2 (since each shortest path found twice, once in each direction)

More detailed example for edge (D,E):
For each root node, we report the value computed for edge (D,E) for Girvan Newman:
| root node | value for (D,E) |
|---|---|
| A | 1 |
| B | 1 |
| C | 1 |
| D | 1 |
| E | 4.5 |
| F | 0 |
| G | .5 |
| total | 9 |
We then divide by 2 to get the final betweenness of (D,E), 4.5.
The reason the value is 1 when the root node is one of A,B,C is that E will be a leaf node for each of these. (Shortest paths to F or G from A,B,C will never traverse edge (D,E)). The value is 0.5 for G as source, becasue half credit is given as there are two shortest paths of length 2 from G to E, but only one traverses (D,E). No other shortest path from root G traverses (D,E).
nx.edge_betweenness_centrality(graph, normalized=False)
{('A', 'B'): 5.0,
('A', 'C'): 1.0,
('B', 'C'): 5.0,
('B', 'D'): 12.0,
('D', 'E'): 4.5,
('D', 'F'): 4.0,
('D', 'G'): 4.5,
('E', 'F'): 1.5,
('F', 'G'): 1.5}
def girvan_newman(G, depth=0):
""" Recursive implementation of the girvan_newman algorithm.
See http://www-rohan.sdsu.edu/~gawron/python_for_ss/course_core/book_draft/Social_Networks/Networkx.html
Args:
G.....a networkx graph
Returns:
A list of all discovered communities,
a list of lists of nodes. """
if G.order() == 1:
return [G.nodes()]
def find_best_edge(G0):
eb = nx.edge_betweenness_centrality(G0)
# eb is dict of (edge, score) pairs, where higher is better
# Return the edge with the highest score.
return sorted(eb.items(), key=lambda x: x[1], reverse=True)[0][0]
# Each component is a separate community. We cluster each of these.
components = [c for c in nx.connected_component_subgraphs(G)]
indent = ' ' * depth # for printing
while len(components) == 1:
edge_to_remove = find_best_edge(G)
print(indent + 'removing ' + str(edge_to_remove))
G.remove_edge(*edge_to_remove)
components = [c for c in nx.connected_component_subgraphs(G)]
result = [c.nodes() for c in components]
print(indent + 'components=' + str(result))
for c in components:
result.extend(girvan_newman(c, depth + 1))
return result
result = girvan_newman(create_example_graph())
removing ('B', 'D')
components=[NodeView(('A', 'C', 'B')), NodeView(('D', 'E', 'F', 'G'))]
removing ('A', 'B')
removing ('A', 'C')
components=[NodeView(('A',)), NodeView(('C', 'B'))]
removing ('C', 'B')
components=[NodeView(('C',)), NodeView(('B',))]
removing ('D', 'E')
removing ('E', 'F')
components=[NodeView(('D', 'F', 'G')), NodeView(('E',))]
removing ('D', 'F')
removing ('D', 'G')
components=[NodeView(('D',)), NodeView(('F', 'G'))]
removing ('F', 'G')
components=[NodeView(('F',)), NodeView(('G',))]
result
[NodeView(('A', 'C', 'B')),
NodeView(('D', 'E', 'F', 'G')),
NodeView(('A',)),
NodeView(('C', 'B')),
NodeView(('A',)),
NodeView(('C',)),
NodeView(('B',)),
NodeView(('C',)),
NodeView(('B',)),
NodeView(('D', 'F', 'G')),
NodeView(('E',)),
NodeView(('D',)),
NodeView(('F', 'G')),
NodeView(('D',)),
NodeView(('F',)),
NodeView(('G',)),
NodeView(('F',)),
NodeView(('G',)),
NodeView(('E',))]
from IPython.core.display import HTML
HTML(open('../custom.css').read())