#!/usr/bin/env python
# coding: utf-8
# # Easy strain design using a high-level interface
#
# WARNING: if you're running this notebook on [try.cameo.bio](http://try.cameo.bio), things might run very slow due to our inability to provide access to the [CPLEX](https://www-01.ibm.com/software/commerce/optimization/cplex-optimizer/) solver on a public webserver. Furthermore, Jupyter kernels might crash and restart due to memory limitations on the server. To avoid that, we encourage the users to shutdown previously opened notebooks. You can do from the `Home` page that by selecting the notebooks highlighted in green and pressing the `Shutdown` button on the top of the menu.
#
# Users primarily interested in using cameo as a tool
# for enumerating metabolic engineering strategies have access to cameo's advanced programming interface via `cameo.api`
# that provides access to potential products (`cameo.api.products`), host organisms (`cameo.api.hosts`) and
# a configurable design function (`cameo.api.design`). Running `cameo.api.design` requires only minimal input and will run the following workflow.
# 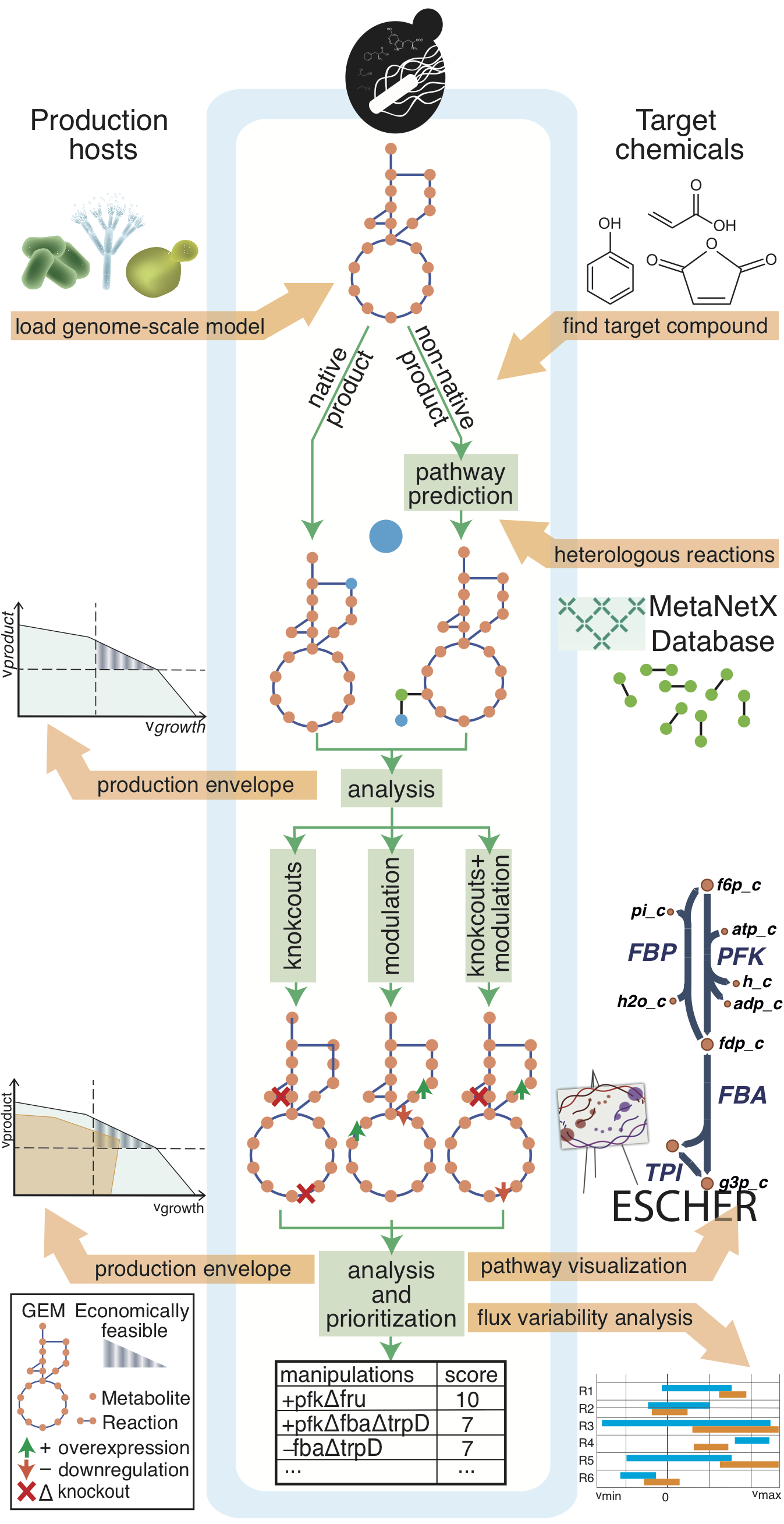 # Import the advanced interface.
# In[1]:
from cameo import api
# ## Searching for products
# Search by trivial name.
# In[2]:
api.products.search('caffeine')
# Search by ChEBI ID.
# In[3]:
api.products.search('chebi:27732')
# ## Host organisms
# Currently the following host organisms and respective models are available in cameo. More hosts and models will be added in the future (please get in touch with us if you'd like to get a particular host organism included).
# In[4]:
for host in api.hosts:
for model in host.models:
print(host.name, model.id)
# ## Computing strain engineering strategies
# For demonstration purposes, we'll set a few options to limit the computational time. Also we'll create a multiprocessing view to take advantage of multicore CPUs (strain design algorithms will be run in parallel for individually predicted heterologous pathways).
# In[5]:
from cameo.parallel import MultiprocessingView
mp_view = MultiprocessingView()
# Limit the number of predicted heterlogous pathways to 4.
# In[6]:
api.design.options.max_pathway_predictions = 4
# Set a time limit of 30 minutes on individual heuristic optimizations.
# In[7]:
api.design.options.heuristic_optimization_timeout = 30
# In[8]:
report = api.design(product='vanillin', view=mp_view)
# In[9]:
report
# ### IPython notebook
# Click [here](http://nbviewer.ipython.org/github/biosustain/cameo/blob/devel/docs/cameo_high_level_interface.ipynb) to download this page as an IPython notebook.
# Import the advanced interface.
# In[1]:
from cameo import api
# ## Searching for products
# Search by trivial name.
# In[2]:
api.products.search('caffeine')
# Search by ChEBI ID.
# In[3]:
api.products.search('chebi:27732')
# ## Host organisms
# Currently the following host organisms and respective models are available in cameo. More hosts and models will be added in the future (please get in touch with us if you'd like to get a particular host organism included).
# In[4]:
for host in api.hosts:
for model in host.models:
print(host.name, model.id)
# ## Computing strain engineering strategies
# For demonstration purposes, we'll set a few options to limit the computational time. Also we'll create a multiprocessing view to take advantage of multicore CPUs (strain design algorithms will be run in parallel for individually predicted heterologous pathways).
# In[5]:
from cameo.parallel import MultiprocessingView
mp_view = MultiprocessingView()
# Limit the number of predicted heterlogous pathways to 4.
# In[6]:
api.design.options.max_pathway_predictions = 4
# Set a time limit of 30 minutes on individual heuristic optimizations.
# In[7]:
api.design.options.heuristic_optimization_timeout = 30
# In[8]:
report = api.design(product='vanillin', view=mp_view)
# In[9]:
report
# ### IPython notebook
# Click [here](http://nbviewer.ipython.org/github/biosustain/cameo/blob/devel/docs/cameo_high_level_interface.ipynb) to download this page as an IPython notebook.
 # Import the advanced interface.
# In[1]:
from cameo import api
# ## Searching for products
# Search by trivial name.
# In[2]:
api.products.search('caffeine')
# Search by ChEBI ID.
# In[3]:
api.products.search('chebi:27732')
# ## Host organisms
# Currently the following host organisms and respective models are available in cameo. More hosts and models will be added in the future (please get in touch with us if you'd like to get a particular host organism included).
# In[4]:
for host in api.hosts:
for model in host.models:
print(host.name, model.id)
# ## Computing strain engineering strategies
# For demonstration purposes, we'll set a few options to limit the computational time. Also we'll create a multiprocessing view to take advantage of multicore CPUs (strain design algorithms will be run in parallel for individually predicted heterologous pathways).
# In[5]:
from cameo.parallel import MultiprocessingView
mp_view = MultiprocessingView()
# Limit the number of predicted heterlogous pathways to 4.
# In[6]:
api.design.options.max_pathway_predictions = 4
# Set a time limit of 30 minutes on individual heuristic optimizations.
# In[7]:
api.design.options.heuristic_optimization_timeout = 30
# In[8]:
report = api.design(product='vanillin', view=mp_view)
# In[9]:
report
# ### IPython notebook
# Click [here](http://nbviewer.ipython.org/github/biosustain/cameo/blob/devel/docs/cameo_high_level_interface.ipynb) to download this page as an IPython notebook.
# Import the advanced interface.
# In[1]:
from cameo import api
# ## Searching for products
# Search by trivial name.
# In[2]:
api.products.search('caffeine')
# Search by ChEBI ID.
# In[3]:
api.products.search('chebi:27732')
# ## Host organisms
# Currently the following host organisms and respective models are available in cameo. More hosts and models will be added in the future (please get in touch with us if you'd like to get a particular host organism included).
# In[4]:
for host in api.hosts:
for model in host.models:
print(host.name, model.id)
# ## Computing strain engineering strategies
# For demonstration purposes, we'll set a few options to limit the computational time. Also we'll create a multiprocessing view to take advantage of multicore CPUs (strain design algorithms will be run in parallel for individually predicted heterologous pathways).
# In[5]:
from cameo.parallel import MultiprocessingView
mp_view = MultiprocessingView()
# Limit the number of predicted heterlogous pathways to 4.
# In[6]:
api.design.options.max_pathway_predictions = 4
# Set a time limit of 30 minutes on individual heuristic optimizations.
# In[7]:
api.design.options.heuristic_optimization_timeout = 30
# In[8]:
report = api.design(product='vanillin', view=mp_view)
# In[9]:
report
# ### IPython notebook
# Click [here](http://nbviewer.ipython.org/github/biosustain/cameo/blob/devel/docs/cameo_high_level_interface.ipynb) to download this page as an IPython notebook.